8WAQ
 
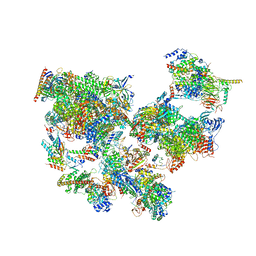 | | Structure of transcribing complex 7 (TC7), the initially transcribing complex with Pol II positioned 7nt downstream of TSS. | | 分子名称: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | 著者 | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | 登録日 | 2023-09-08 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (6.29 Å) | | 主引用文献 | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8U4S
 
 | |
8U4R
 
 | |
8YB5
 
 | |
2JKT
 
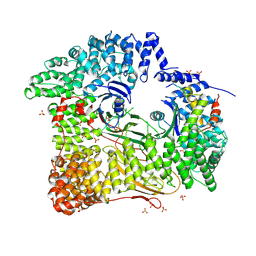 | | AP2 CLATHRIN ADAPTOR CORE with CD4 Dileucine peptide RM(phosphoS) EIKRLLSE Q to E mutant | | 分子名称: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA-1, AP-2 COMPLEX SUBUNIT MU-1, ... | | 著者 | Owen, D.J, McCoy, A.J, Kelly, B.T, Evans, P.R. | | 登録日 | 2008-08-29 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | A Structural Explanation for the Binding of Endocytic Dileucine Motifs by the Ap2 Complex.
Nature, 456, 2008
|
|
2JKR
 
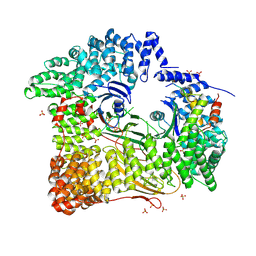 | | AP2 CLATHRIN ADAPTOR CORE with Dileucine peptide RM(phosphoS)QIKRLLSE | | 分子名称: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA-1, AP-2 COMPLEX SUBUNIT MU-1, ... | | 著者 | Owen, D.J, McCoy, A.J, Kelly, B.T, Evans, P.R. | | 登録日 | 2008-08-29 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | A Structural Explanation for the Binding of Endocytic Dileucine Motifs by the Ap2 Complex.
Nature, 456, 2008
|
|
2KA4
 
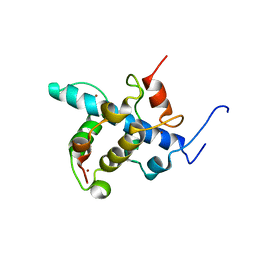 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | 分子名称: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | 著者 | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | 登録日 | 2008-10-30 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2K05
 
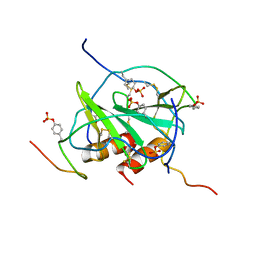 | |
2KA6
 
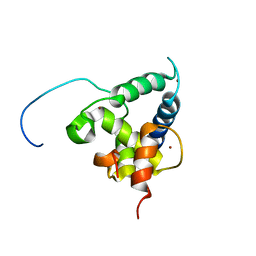 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | 分子名称: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | 著者 | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | 登録日 | 2008-10-30 | | 公開日 | 2009-04-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
2KQV
 
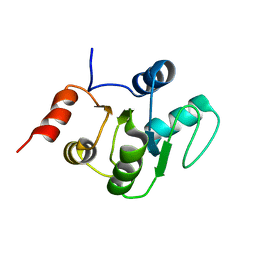 | |
2I2X
 
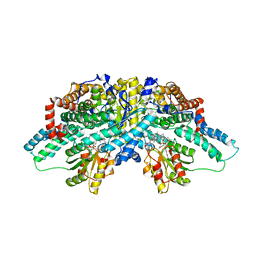 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | 分子名称: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | 著者 | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | 登録日 | 2006-08-17 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2KQW
 
 | |
2K2U
 
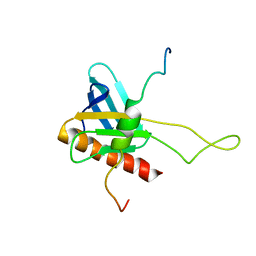 | | NMR Structure of the complex between Tfb1 subunit of TFIIH and the activation domain of VP16 | | 分子名称: | Alpha trans-inducing protein, RNA polymerase II transcription factor B subunit 1 | | 著者 | Langlois, C, Mas, C, Di Lello, P, Miller Jenkins, P.M, Legault, J, Omichinski, J.G. | | 登録日 | 2008-04-11 | | 公開日 | 2008-08-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the Complex between the Tfb1 Subunit of TFIIH and the Activation Domain of VP16: Structural Similarities between VP16 and p53.
J.Am.Chem.Soc., 130, 2008
|
|
2K04
 
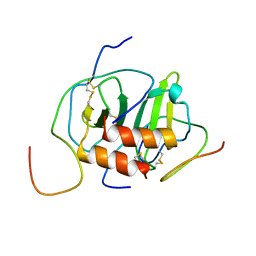 | |
2K03
 
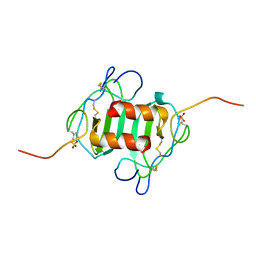 | |
3GHG
 
 | | Crystal Structure of Human Fibrinogen | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A knob, B knob, ... | | 著者 | Doolittle, R.F, Kollman, J.M, Sawaya, M.R, Pandi, L, Riley, M. | | 登録日 | 2009-03-03 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-05-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of human fibrinogen.
Biochemistry, 48, 2009
|
|
3GCM
 
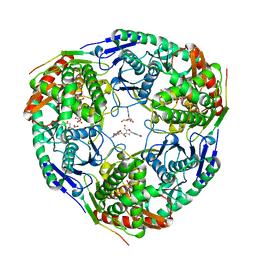 | |
3GME
 
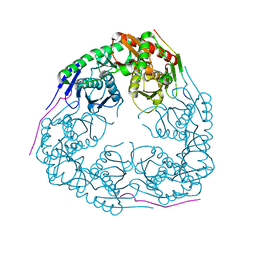 | |
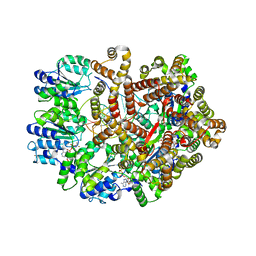
3GLF
 
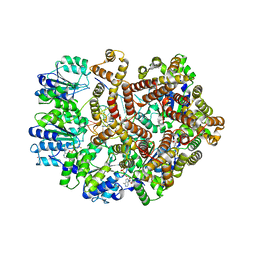 | |
3GLG
 
 | | Crystal Structure of a Mutant (gammaT157A) E. coli Clamp Loader Bound to Primer-Template DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | 著者 | Simonetta, K.R, Seyedin, S.N, Kuriyan, J. | | 登録日 | 2009-03-12 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GYX
 
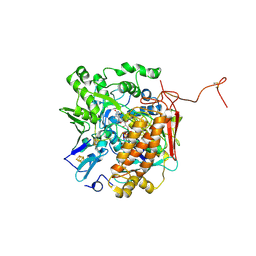 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | 分子名称: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | 著者 | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | 登録日 | 2009-04-06 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
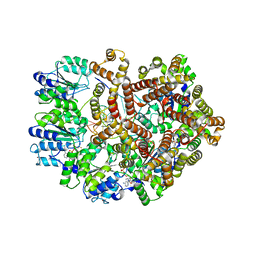
3GLI
 
 | | Crystal Structure of the E. coli clamp loader bound to Primer-Template DNA and Psi Peptide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | 著者 | Simonetta, K.R, Cantor, A.J, Kuriyan, J. | | 登録日 | 2009-03-12 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
3H1C
 
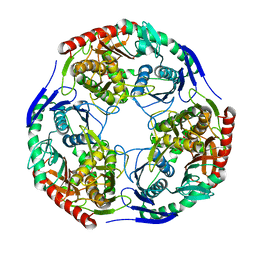 | |
3GLH
 
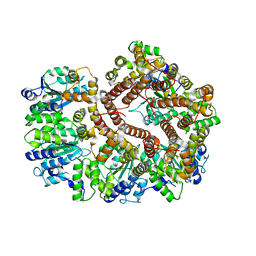 | | Crystal Structure of the E. coli clamp loader bound to Psi Peptide | | 分子名称: | DNA polymerase III subunit delta, DNA polymerase III subunit delta', DNA polymerase III subunit tau | | 著者 | Kazmirski, S.L, Simonetta, K.R, Kuriyan, J. | | 登録日 | 2009-03-12 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.891 Å) | | 主引用文献 | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | 登録日 | 2022-09-21 | | 公開日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (5.04 Å) | | 主引用文献 | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
