6YI1
 
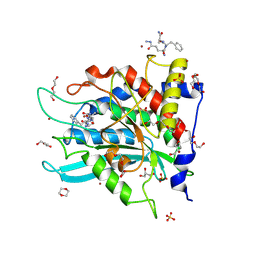 | | Crystal structure of human glutaminyl cyclase in complex with Glu(gamma-hydrazide)-Phe-Ala | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kupski, O, Sautner, V, Tittmann, K. | | 登録日 | 2020-03-31 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Hydrazides Are Potent Transition-State Analogues for Glutaminyl Cyclase Implicated in the Pathogenesis of Alzheimer's Disease.
Biochemistry, 59, 2020
|
|
6YJN
 
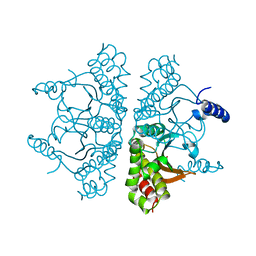 | |
7O2M
 
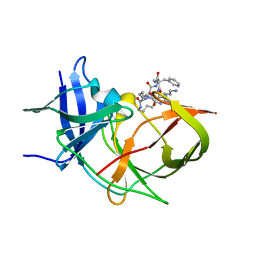 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2289 | | 分子名称: | 1-[(3~{S},6~{S},9~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-9-(phenylmethyl)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, Genome polyprotein | | 著者 | Huber, S, Heine, A, Steinmetzer, T. | | 登録日 | 2021-03-30 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7OC2
 
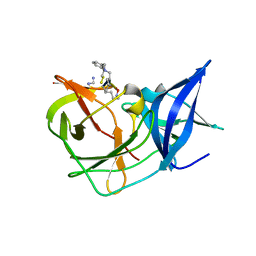 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2295 | | 分子名称: | Cyclic 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE-(7-3)-7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE-(7-19)-N-ACETYL-L-CYSTEINE-(8-25)-[3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID-()-(6E,11E)-HEPTADECA-6,11-DIENE-9,9-DIYLBIS(PHOSPHONIC ACID), Serine protease NS3, Serine protease subunit NS2B | | 著者 | Huber, S, Heine, A, Steinmetzer, T. | | 登録日 | 2021-04-25 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
6YQ9
 
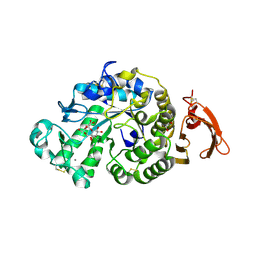 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | 分子名称: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | 登録日 | 2020-04-16 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
7O55
 
 | |
2CFZ
 
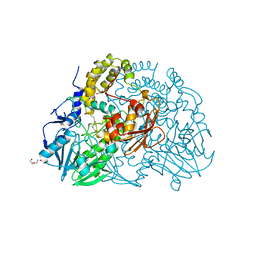 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-dodecanol | | 分子名称: | 1-DODECANOL, DI(HYDROXYETHYL)ETHER, SDS HYDROLASE SDSA1, ... | | 著者 | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | 登録日 | 2006-02-26 | | 公開日 | 2006-04-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6YQC
 
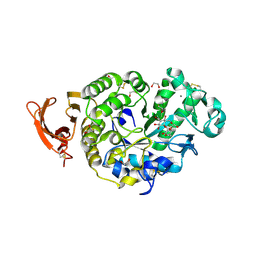 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | 分子名称: | (1~{R},2~{S},4~{R},5~{S},6~{R})-6-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-heptoxy-6-(hydroxymethyl)-3,4-bis(oxidanyl)oxan-2-yl]oxy-5-(hydroxymethyl)cyclohexane-1,2,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | 登録日 | 2020-04-16 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
7OBV
 
 | |
6YQB
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol cyclosulfate inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | 著者 | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | 登録日 | 2020-04-16 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
5JOM
 
 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | 登録日 | 2016-05-02 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2CG2
 
 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with sulfate | | 分子名称: | SDSA1, SULFATE ION, ZINC ION | | 著者 | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | 登録日 | 2006-02-27 | | 公開日 | 2006-04-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CLD
 
 | |
6YQA
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol aziridine inhibitor | | 分子名称: | (1~{S},2~{R},3~{R},4~{R},5~{R})-5-(8-azanyloctylamino)-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | 登録日 | 2020-04-16 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
2CL7
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A FLUORESCENT FORM OF H-RAS P21 IN COMPLEX WITH GTP | | 分子名称: | GTPASE HRAS, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Klink, B.U, Goody, R.S, Scheidig, A.J. | | 登録日 | 2006-04-26 | | 公開日 | 2006-05-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A Newly Designed Microspectrofluorometer for Kinetic Studies on Protein Crystals in Combination with X-Ray Diffraction.
Biophys.J., 91, 2006
|
|
5UAD
 
 | |
1A1S
 
 | | ORNITHINE CARBAMOYLTRANSFERASE FROM PYROCOCCUS FURIOSUS | | 分子名称: | ORNITHINE CARBAMOYLTRANSFERASE | | 著者 | Villeret, V, Clantin, B, Tricot, C, Legrain, C, Roovers, M, Stalon, V, Glansdorff, N, Van Beeumen, J. | | 登録日 | 1997-12-15 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2CI4
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I crystal form II | | 分子名称: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | 著者 | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | 登録日 | 2006-03-17 | | 公開日 | 2006-05-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
5K5E
 
 | | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | De la Mora, E, Dighe, S.N, Deora, G.S, Ross, B.P, Nachon, F, Brazzolotto, X. | | 登録日 | 2016-05-23 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening.
J.Med.Chem., 59, 2016
|
|
8AEB
 
 | | SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(pyridin-3-ylmethyl)thioformamide, ... | | 著者 | Hanoulle, X, Charton, J, Deprez, B. | | 登録日 | 2022-07-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
1TV2
 
 | | Crystal structure of the hydroxylamine MtmB complex | | 分子名称: | 5-HYDROXYAMINO-3-METHYL-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1 | | 著者 | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | 登録日 | 2004-06-26 | | 公開日 | 2004-10-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
7O70
 
 | | KRasG12C ligand complex | | 分子名称: | 1-[(4R,7S)-12-chloro-14-fluoro-13-(2-fluoro-6-hydroxyphenyl)-4-methyl-10-oxa-2,5,16,18-tetrazatetracyclo[9.7.1.0^(2,7).0^(15,19)]nonadeca-1(18),11,13,15(19),16-pentaen-5-en-1-one-yl]prop-2, CALCIUM ION, GTPase KRas, ... | | 著者 | Phillips, C. | | 登録日 | 2021-04-12 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
2CI3
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase crystal form I | | 分子名称: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | 著者 | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | 登録日 | 2006-03-17 | | 公開日 | 2006-05-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
7O83
 
 | | KRasG12C ligand complex | | 分子名称: | 1-[(7S)-11-chloro-12-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,15,17-tetrazatetracyclo[8.7.1.02,7.014,18]octadeca-1(17),10,12,14(18),15-pentaen-5-yl]prop-2-en-1-one, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Phillips, C, Breed, J. | | 登録日 | 2021-04-14 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
6YO6
 
 | | Structure of iC3b1 | | 分子名称: | hC3Nb1, iC3b1 alpha chain, iC3b1 beta chain | | 著者 | Jensen, R.K, Andersen, G.R. | | 登録日 | 2020-04-14 | | 公開日 | 2021-02-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Complement Receptor 3 Forms a Compact High-Affinity Complex with iC3b.
J Immunol., 206, 2021
|
|
