1K79
 
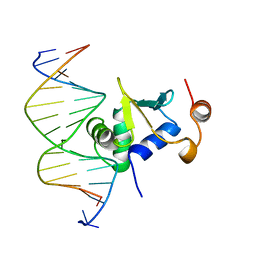 | | Ets-1(331-440)+GGAA duplex | | 分子名称: | C-ets-1 protein, DNA (5'-D(*CP*AP*CP*AP*TP*TP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*AP*AP*TP*GP*T)-3') | | 著者 | Garvie, C.W, Hagman, J, Wolberger, C. | | 登録日 | 2001-10-18 | | 公開日 | 2002-01-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
2K05
 
 | |
2N55
 
 | |
2K04
 
 | |
1K78
 
 | |
2K03
 
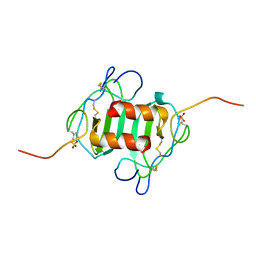 | |
1H8G
 
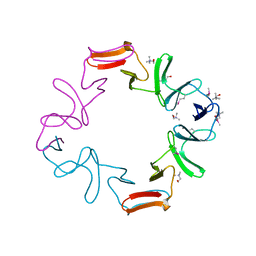 | | C-terminal domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | 分子名称: | CHOLINE ION, MAJOR AUTOLYSIN | | 著者 | Fernandez-Tornero, C, Garcia, E, Lopez, R, Gimenez-Gallego, G, Romero, A. | | 登録日 | 2001-02-06 | | 公開日 | 2002-01-31 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
2FPF
 
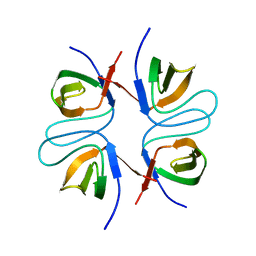 | |
2FPD
 
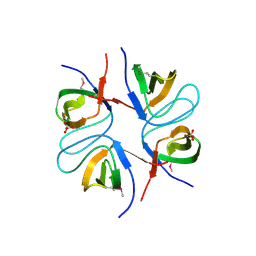 | |
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2002-02-05 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
4VHB
 
 | | THIOCYANATE ADDUCT OF THE BACTERIAL HEMOGLOBIN FROM VITREOSCILLA SP. | | 分子名称: | PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | 著者 | Bolognesi, M, Boffi, A, Coletta, M, Mozzarelli, A, Pesce, A, Tarricone, C, Ascenzi, P. | | 登録日 | 1999-03-11 | | 公開日 | 1999-08-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
J.Mol.Biol., 291, 1999
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | 分子名称: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | 著者 | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | 登録日 | 2014-08-08 | | 公開日 | 2015-08-05 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
6ZGN
 
 | | Crystal structure of VirB8-like OrfG central domain of Streptococcus thermophilus ICESt3; a putative assembly factor of a gram positive conjugative Type IV secretion system. | | 分子名称: | Putative transfer protein | | 著者 | Cappele, J, Mohamad-Ali, A, Leblond-Bourget, N, Payot-Lacroix, S, Mathiot, S, Didierjean, C, Favier, F, Douzi, B. | | 登録日 | 2020-06-19 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and Biochemical Analysis of OrfG: The VirB8-like Component of the Conjugative Type IV Secretion System of ICE St3 From Streptococcus thermophilus .
Front Mol Biosci, 8, 2021
|
|
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | 分子名称: | TRYPSIN INHIBITOR II | | 著者 | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | 登録日 | 2001-04-02 | | 公開日 | 2001-04-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
5J7T
 
 | |
4V9G
 
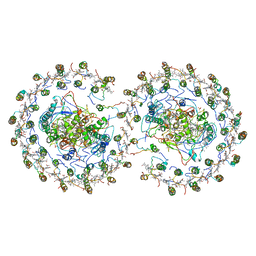 | | RC-LH1-PufX dimer complex from Rhodobacter sphaeroides | | 分子名称: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | 著者 | Qian, P, Papiz, M.Z, Jackson, P.J, Brindley, A.A, Ng, I.W, Olsen, J.D, Dickman, M.J, Bullough, P.A, Hunter, C.N. | | 登録日 | 2013-02-21 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (7.78 Å) | | 主引用文献 | Three-Dimensional Structure of the Rhodobacter sphaeroides RC-LH1-PufX Complex: Dimerization and Quinone Channels Promoted by PufX.
Biochemistry, 52, 2013
|
|
5IW6
 
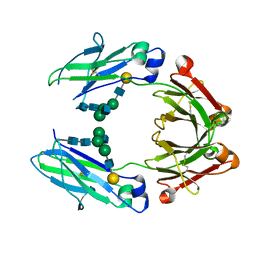 | | anti-CD20 monoclonal antibody Fc fragment | | 分子名称: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Tang, C, Chen, Z. | | 登録日 | 2016-03-22 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure of anti-CD20 monoclonal antibody Fc fragment at 2.34 Angstroms resolution
To Be Published
|
|
4WCJ
 
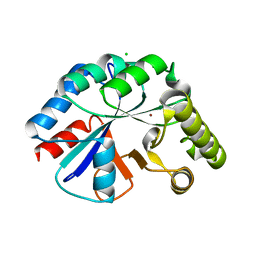 | | Structure of IcaB from Ammonifex degensii | | 分子名称: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | 著者 | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | 登録日 | 2014-09-04 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
1HUI
 
 | | INSULIN MUTANT (B1, B10, B16, B27)GLU, DES-B30, NMR, 25 STRUCTURES | | 分子名称: | INSULIN | | 著者 | Olsen, H.B, Ludvigsen, S, Kaarsholm, N.C. | | 登録日 | 1996-03-29 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an engineered insulin monomer at neutral pH.
Biochemistry, 35, 1996
|
|
3BLG
 
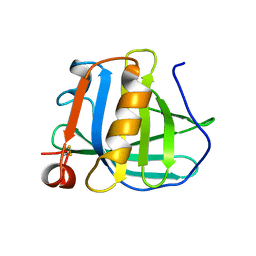 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 6.2 | | 分子名称: | BETA-LACTOGLOBULIN | | 著者 | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | 登録日 | 1998-08-29 | | 公開日 | 1999-01-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
3BZC
 
 | |
3BZK
 
 | |
5IUF
 
 | |
2Q34
 
 | |
5J03
 
 | |
