3RPV
 
 | | CDK2 in complex with inhibitor RC-2-88 | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(3-aminobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-04-27 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3R1V
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, in complex with an azo compound | | 分子名称: | 4-{(E)-[4-(propan-2-yl)phenyl]diazenyl}phenol, Odorant binding protein, antennal | | 著者 | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | 登録日 | 2011-03-11 | | 公開日 | 2011-10-19 | | 最終更新日 | 2011-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3RUN
 
 | | New strategy to analyze structures of glycopeptide antibiotic-target complexes | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Economou, N.J, Townsend, T.M, Loll, P.J. | | 登録日 | 2011-05-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A carrier protein strategy yields the structure of dalbavancin.
J.Am.Chem.Soc., 134, 2012
|
|
3SU5
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with vaniprevir | | 分子名称: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, NS4A protein, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2011-07-11 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU0
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with danoprevir | | 分子名称: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2011-07-11 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.159 Å) | | 主引用文献 | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SMQ
 
 | | Crystal structure of protein arginine methyltransferase 3 | | 分子名称: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | 著者 | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | 登録日 | 2011-06-28 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
3RNI
 
 | | CDK2 in complex with inhibitor RC-3-86 | | 分子名称: | 3-[(4-amino-5-benzoyl-1,3-thiazol-2-yl)amino]benzenesulfonamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-04-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RAL
 
 | | CDK2 in complex with inhibitor RC-2-34 | | 分子名称: | 4-{[4-amino-5-(3-methoxybenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RD9
 
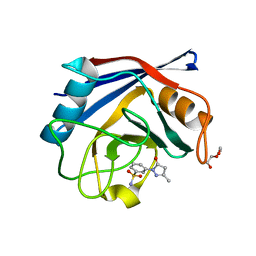 | | Human Cyclophilin D Complexed with a Fragment | | 分子名称: | 3-(3-methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl)benzenesulfonamide, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase F, ... | | 著者 | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | 登録日 | 2011-04-01 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
3D76
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, soaked at pH 7.0 | | 分子名称: | 1,2-ETHANEDIOL, N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1, ... | | 著者 | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | 登録日 | 2008-05-20 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
3RUM
 
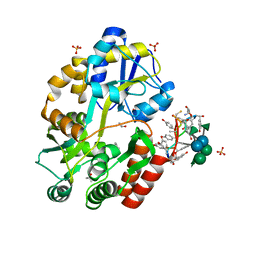 | | New strategy to analyze structures of glycopeptide antibiotic-target complexes | | 分子名称: | 3-amino-2,3,6-trideoxy-alpha-L-ribo-hexopyranose, ISOPROPYL ALCOHOL, Maltose-binding periplasmic protein, ... | | 著者 | Economou, N.J, Weeks, S.D, Grasty, K.C, Nahoum, V, Loll, P.J. | | 登録日 | 2011-05-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | A carrier protein strategy yields the structure of dalbavancin.
J.Am.Chem.Soc., 134, 2012
|
|
3RUL
 
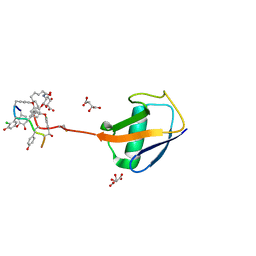 | | New strategy to analyze structures of glycopeptide-target complexes | | 分子名称: | 10-METHYLUNDECANOIC ACID, 2-amino-2-deoxy-beta-D-glucopyranuronic acid, CHLORIDE ION, ... | | 著者 | Economou, N.J, Nahoum, V, Weeks, S.D, Grasty, K.C, Loll, P.J. | | 登録日 | 2011-05-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A carrier protein strategy yields the structure of dalbavancin.
J.Am.Chem.Soc., 134, 2012
|
|
3S1H
 
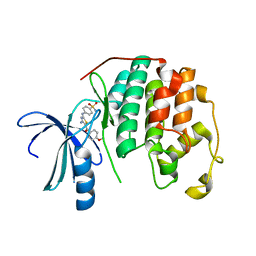 | | CDK2 in complex with inhibitor RC-2-39 | | 分子名称: | 4-{[4-amino-5-(4-methoxybenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-05-15 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3SK2
 
 | |
3SKE
 
 | | I. Novel HCV NS5B Polymerase Inhibitors: Discovery of Indole 2- Carboxylic Acids with C3-Heterocycles | | 分子名称: | 1-[(2-aminopyridin-4-yl)methyl]-3-(2,4-dioxo-1,2-dihydrothieno[3,4-d]pyrimidin-3(4H)-yl)-5-(trifluoromethyl)-1H-indole-2-carboxylic acid, HCV NS5B RNA_DEPENDENT RNA POLYMERASE, PHOSPHATE ION | | 著者 | Lesburg, C.A, Anilkumar, G.N. | | 登録日 | 2011-06-22 | | 公開日 | 2011-08-31 | | 最終更新日 | 2012-09-26 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | I. Novel HCV NS5B polymerase inhibitors: discovery of indole 2-carboxylic acids with C3-heterocycles.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3S3K
 
 | |
3SK1
 
 | |
3SU3
 
 | | Crystal structure of NS3/4A protease in complex with vaniprevir | | 分子名称: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, NS4A protein, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2011-07-11 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUG
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with MK-5172 | | 分子名称: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2011-07-11 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SV9
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2011-07-12 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SSA
 
 | | Crystal structure of subunit B mutant N157T of the A1AO ATP synthase | | 分子名称: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sundararaman, L, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2011-07-08 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
3RAK
 
 | | CDK2 in complex with inhibitor RC-2-32 | | 分子名称: | 4-[(4-amino-5-benzoyl-1,3-thiazol-2-yl)amino]benzenesulfonamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3SKH
 
 | |
3STO
 
 | | Serpin from the trematode Schistosoma Haematobium | | 分子名称: | Serine protease inhibitor | | 著者 | Granzin, J, Weiergraeber, O.H, Lee, X, Blanton, R.E. | | 登録日 | 2011-07-11 | | 公開日 | 2012-05-30 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Three-dimensional structure of a schistosome serpin revealing an unusual configuration of the helical subdomain.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SUE
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | 分子名称: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2011-07-11 | | 公開日 | 2012-09-19 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
