1L46
 
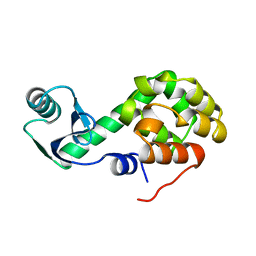 | |
1L42
 
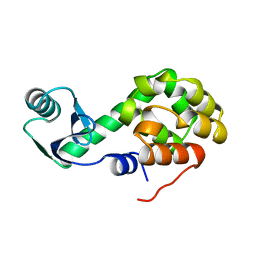 | |
1L54
 
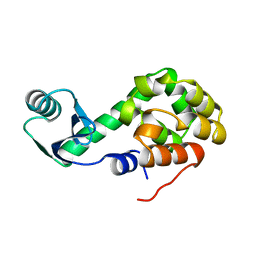 | |
1EQT
 
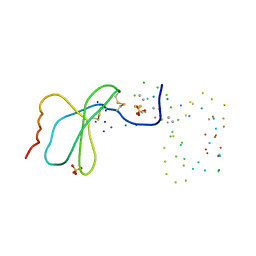 | | MET-RANTES | | 分子名称: | SULFATE ION, T-CELL SPECIFIC RANTES PROTEIN | | 著者 | Hoover, D.M, Shaw, J, Gryczynski, Z, Proudfoot, A.E.I, Wells, T. | | 登録日 | 2000-04-06 | | 公開日 | 2000-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Crystal Structure of MET-RANTES: Comparison with Native RANTES and AOP-RANTES
PROTEIN PEPT.LETT., 7, 2000
|
|
6TWA
 
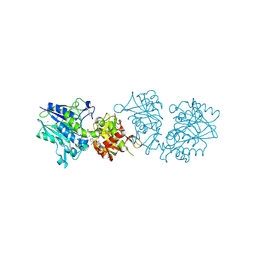 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12646 (an AOPCP derivative, compound 20 in publication) in the closed state | | 分子名称: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | 著者 | Pippel, J, Strater, N. | | 登録日 | 2020-01-12 | | 公開日 | 2020-02-19 | | 最終更新日 | 2020-04-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TWF
 
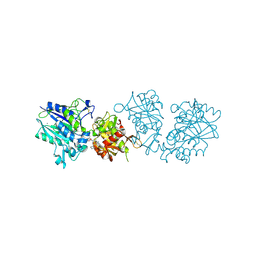 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12604 (an AOPCP derivative, compound 21 in publication) in the closed state | | 分子名称: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | 著者 | Pippel, J, Strater, N. | | 登録日 | 2020-01-13 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TW0
 
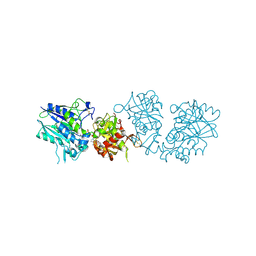 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12690 (an AOPCP derivative, compound 10 in publication) in the closed state | | 分子名称: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-2-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | 著者 | Pippel, J, Strater, N. | | 登録日 | 2020-01-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TVX
 
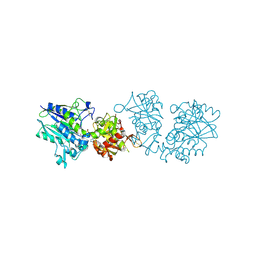 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12676 (an AOPCP derivative, compound 9 in paper) in the closed state | | 分子名称: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | 著者 | Pippel, J, Strater, N. | | 登録日 | 2020-01-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
3WXX
 
 | | Crystal Structure of a T3SS complex from Aeromonas hydrophila | | 分子名称: | AcrH, AopB, MAGNESIUM ION | | 著者 | Nguyen, V.S, Jobichen, C, Sivaraman, J, Henry, Y.K.M. | | 登録日 | 2014-08-12 | | 公開日 | 2015-10-14 | | 最終更新日 | 2015-11-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of AcrH-AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly
Structure, 23, 2015
|
|
4AOP
 
 | |
7AOP
 
 | | Structure of NUDT15 in complex with inhibitor TH8321 | | 分子名称: | 2-azanyl-9-cyclohexyl-8-(2-methoxyphenyl)-3~{H}-purine-6-thione, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | 著者 | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | 登録日 | 2020-10-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
2AOP
 
 | | SULFITE REDUCTASE: REDUCED WITH CRII EDTA, SIROHEME FEII, [4FE-4S] +1, PHOSPHATE BOUND | | 分子名称: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-09 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
3AOP
 
 | | SULFITE REDUCTASE HEMOPROTEIN PHOTOREDUCED WITH PROFLAVINE EDTA, SIROHEME FEII,[4FE-4S] +1, PHOSPHATE BOUND | | 分子名称: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-09 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
1AOP
 
 | |
5AOP
 
 | | SULFITE REDUCTASE STRUCTURE REDUCED WITH CRII EDTA, 5-COORDINATE SIROHEME, SIROHEME FEII, [4FE-4S] +1 | | 分子名称: | IRON/SULFUR CLUSTER, POTASSIUM ION, SIROHEME, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-10 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange.
Biochemistry, 36, 1997
|
|
6AOP
 
 | |
8AOP
 
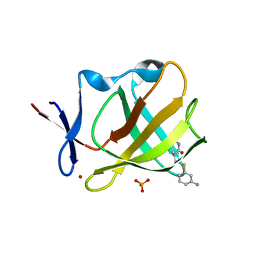 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 14r | | 分子名称: | (3S)-3-[(3-aminophenyl)sulfanylmethyl]piperidine-2,6-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | 著者 | Maiwald, S, Heim, C, Hartmann, M.D. | | 登録日 | 2022-08-08 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.944 Å) | | 主引用文献 | Synthesis of novel glutarimide ligands for the E3 ligase substrate receptor Cereblon (CRBN): Investigation of their binding mode and antiproliferative effects against myeloma cell lines.
Eur.J.Med.Chem., 246, 2023
|
|
1H4O
 
 | |
4G38
 
 | |
4G39
 
 | |
7QDA
 
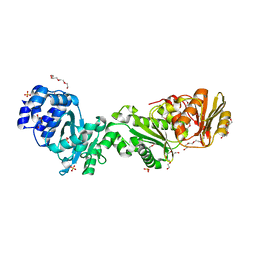 | | Crystal structure of CalpL | | 分子名称: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Schneberger, N, Hagelueken, G. | | 登録日 | 2021-11-26 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
4X5S
 
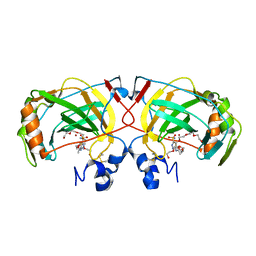 | | The crystal structure of an alpha carbonic anhydrase from the extremophilic bacterium Sulfurihydrogenibium azorense. | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase (Carbonate dehydratase), ... | | 著者 | De Simone, G, Alterio, V, Di Fiore, A. | | 登録日 | 2014-12-05 | | 公開日 | 2015-05-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6K1G
 
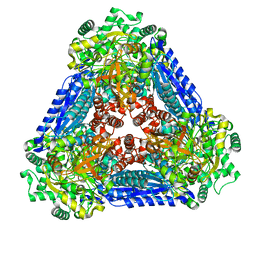 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | 分子名称: | L-fucose isomerase, MANGANESE (II) ION | | 著者 | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | 登録日 | 2019-05-10 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
6K1F
 
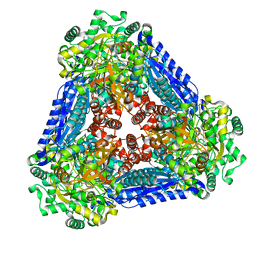 | | Crystal structure of the L-fucose isomerase from Raoultella sp. | | 分子名称: | L-fucose isomerase, MANGANESE (II) ION | | 著者 | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | 登録日 | 2019-05-10 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
2DU5
 
 | |
