4U45
 
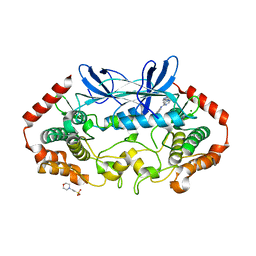 | | MAP4K4 in complex with inhibitor (compound 25) | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(1H-pyrazol-4-yl)-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ... | | 著者 | Harris, S.F, Wu, P, Coons, M. | | 登録日 | 2014-07-23 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5KV4
 
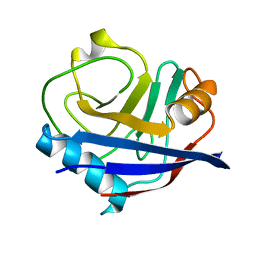 | | Human cyclophilin A at 278K, Data set 6 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | 登録日 | 2016-07-13 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
8SBM
 
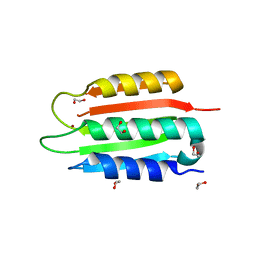 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | 分子名称: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | 著者 | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | 登録日 | 2023-04-03 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
5KVH
 
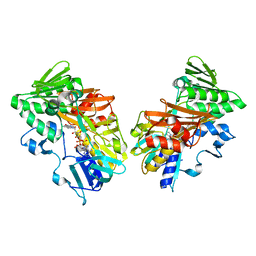 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | 分子名称: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | 登録日 | 2016-07-14 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.273 Å) | | 主引用文献 | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
8SWZ
 
 | | PARP4 ART domain bound to EB47 | | 分子名称: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | 著者 | Frigon, L, Pascal, J.M. | | 登録日 | 2023-05-19 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SLZ
 
 | |
5L1W
 
 | |
7C3I
 
 | | Structure of L-lysine oxidase D212A/D315A | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | 登録日 | 2020-05-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
8SX1
 
 | | PARP4 catalytic domain | | 分子名称: | Protein mono-ADP-ribosyltransferase PARP4 | | 著者 | Frigon, L, Pascal, J.M. | | 登録日 | 2023-05-19 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
5L2A
 
 | |
8SX2
 
 | | PARP4 catalytic domain bound to EB47 | | 分子名称: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Protein mono-ADP-ribosyltransferase PARP4 | | 著者 | Frigon, L, Pascal, J.M. | | 登録日 | 2023-05-19 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SWY
 
 | | PARP4 ART domain bound to NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | 著者 | Frigon, L, Pascal, J.M. | | 登録日 | 2023-05-19 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
5L3E
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6,7-dimethoxyquinazoline-2,4-diamine, ... | | 著者 | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | 登録日 | 2016-04-10 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
7C67
 
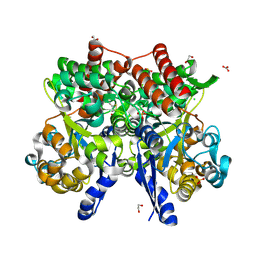 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotriose | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6F
 
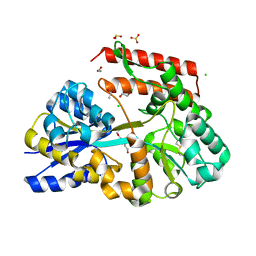 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in an open state | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C71
 
 | | Crystal structure of beta-glycosides-binding protein (E117A) of ABC transporter in an open state | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFITE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-22 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6M
 
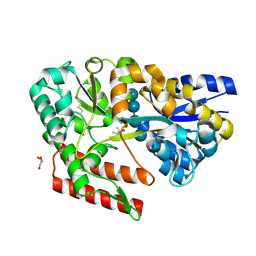 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotetraose (Form I) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
8SBS
 
 | |
7C6Y
 
 | | Crystal structure of beta-glycosides-binding protein (W41A) of ABC transporter in an open state (Form II) | | 分子名称: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | 登録日 | 2020-05-22 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5L7D
 
 | | Structure of human Smoothened in complex with cholesterol | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, SODIUM ION, ... | | 著者 | Byrne, E.F.X, Sircar, R, Miller, P.S, Hedger, G, Luchetti, G, Nachtergaele, S, Tully, M.D, Mydock-McGrane, L, Covey, D.F, Rambo, R.P, Sansom, M.S.P, Newstead, S, Rohatgi, R, Siebold, C. | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of Smoothened regulation by its extracellular domains.
Nature, 535, 2016
|
|
5L8N
 
 | | crystal structure of human FABP6 protein with fragment 1 | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 5,6-dimethyl-1~{H}-benzimidazol-2-amine, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | 登録日 | 2016-06-08 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
7C87
 
 | |
5L8T
 
 | |
8SMD
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-3' gcADPR | | 分子名称: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, ABC transporter ATPase | | 著者 | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | 登録日 | 2023-04-26 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
5L98
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 3-amino-2-methylpyridine derivative 4 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1~{S})-1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethyl]-2-methyl-pyridin-3-amine | | 著者 | Lolli, G, Marchand, J.-R, Caflisch, A. | | 登録日 | 2016-06-09 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.263 Å) | | 主引用文献 | Derivatives of 3-Amino-2-methylpyridine as BAZ2B Bromodomain Ligands: In Silico Discovery and in Crystallo Validation.
J. Med. Chem., 59, 2016
|
|
