6ZN7
 
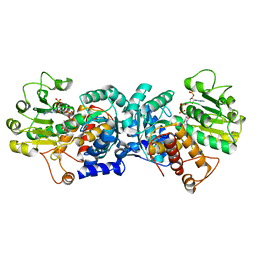 | | MaeB malic enzyme domain apoprotein | | 分子名称: | MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | 著者 | Lovering, A.L, Harding, C.J. | | 登録日 | 2020-07-06 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
9FXU
 
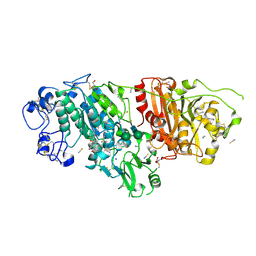 | | Crystal Structure of Autotaxin (ENPP2) with Type VI Inhibitor, a Novel Class of Inhibitors with Three-Point Lock Binding Mode | | 分子名称: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | 著者 | Borza, R, Joosten, R.P, Perrakis, A. | | 登録日 | 2024-07-02 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Design, Synthesis, and Biological Implications of Autotaxin inhibitors with a Three-Point lock binding mode.
Bioorg.Med.Chem., 124, 2025
|
|
5M21
 
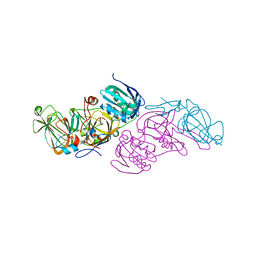 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 with 4-hydroxybenzoate bound | | 分子名称: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | 著者 | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | 登録日 | 2016-10-11 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
6SU9
 
 | |
6ZNU
 
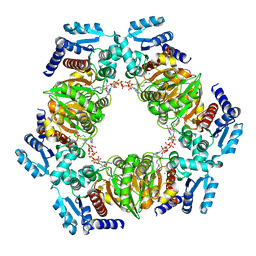 | | MaeB PTA domain E544R mutant | | 分子名称: | ACETYL COENZYME *A, CHLORIDE ION, Malate dehydrogenase | | 著者 | Lovering, A.L, Harding, C.J. | | 登録日 | 2020-07-06 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
7A7K
 
 | |
6SUM
 
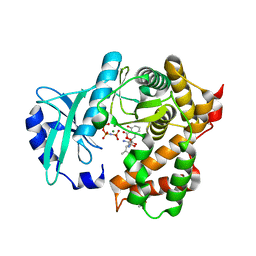 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, MG2+ and Ami | | 分子名称: | ACETATE ION, AMICOUMACIN KINASE, Amicoumacin A, ... | | 著者 | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | 登録日 | 2019-09-16 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUZ
 
 | |
7A80
 
 | |
7A7U
 
 | |
7A87
 
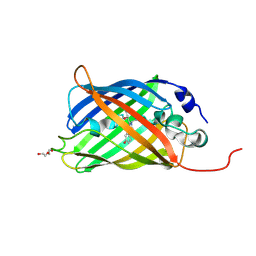 | |
6SW6
 
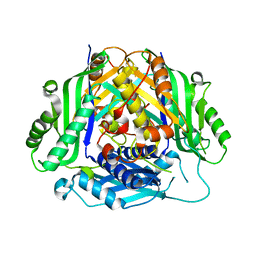 | |
6ZNK
 
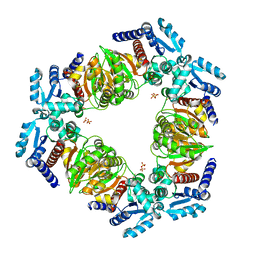 | |
6SWI
 
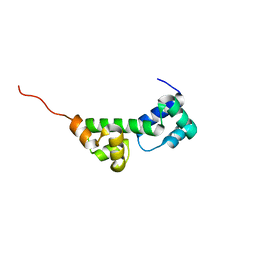 | |
5LDN
 
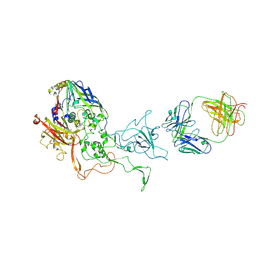 | | A viral capsid:antibody complex | | 分子名称: | Hexon protein,hexon capsid, antibody | | 著者 | James, L. | | 登録日 | 2016-06-27 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Antibody-antigen kinetics constrain intracellular humoral immunity.
Sci Rep, 6, 2016
|
|
6SWL
 
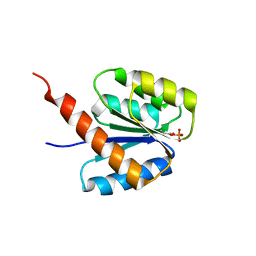 | | The REC domain of XynC, a response regulator from Geobacillus stearothermophilus | | 分子名称: | MAGNESIUM ION, Two-component response regulator | | 著者 | Lansky, S, Lavid, N, Shoham, Y, Shoham, G. | | 登録日 | 2019-09-22 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | The REC domain of XynC, a response regulator from Geobacillus stearothermophilus
To Be Published
|
|
7A82
 
 | |
7A8B
 
 | |
5LH6
 
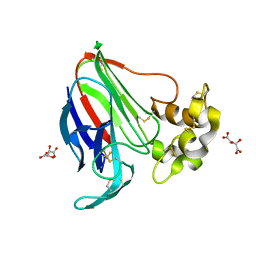 | | High dose Thaumatin - 360-400 ms. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
7A8H
 
 | |
6SLL
 
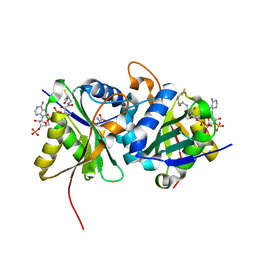 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) and coenzyme A | | 分子名称: | 2,4-DIAMINOBUTYRIC ACID, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase, ... | | 著者 | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | 登録日 | 2019-08-20 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6ZN9
 
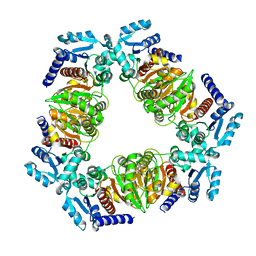 | |
7A8D
 
 | |
5LEX
 
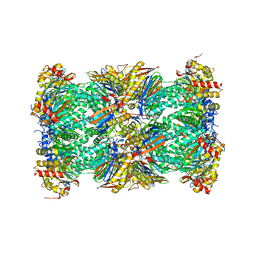 | | Native human 20S proteasome in Mg-Acetate at 2.2 Angstrom | | 分子名称: | MAGNESIUM ION, PENTAETHYLENE GLYCOL, POTASSIUM ION, ... | | 著者 | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | 登録日 | 2016-06-30 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
9FXY
 
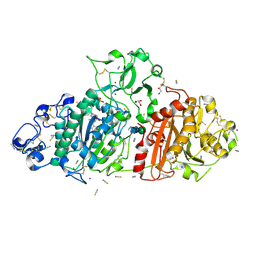 | | Crystal Structure of Autotaxin (ENPP2) with Type IV Inhibitor | | 分子名称: | 3-(3-((4-(4-fluorophenyl)thiazol-2-yl)(methyl)amino)-6-(1-(methylsulfonyl)piperidin-4-yl)imidazo[1,2-b]pyridazin-2-yl)propanenitrile, CALCIUM ION, GLYCEROL, ... | | 著者 | Borza, R, Joosten, R.P, Perrakis, A. | | 登録日 | 2024-07-02 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, Synthesis, and Biological Implications of Autotaxin inhibitors with a Three-Point lock binding mode.
Bioorg.Med.Chem., 124, 2025
|
|
