5SSQ
 
 | |
5SPE
 
 | |
5SOJ
 
 | |
5SPF
 
 | |
5SOQ
 
 | |
5SPG
 
 | |
5SOI
 
 | |
5SPI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4574659604 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPA
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894417 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-(4-carbamamidobenzamido)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPH
 
 | |
5SPB
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894404 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[4-(phenylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPK
 
 | |
5SOL
 
 | |
5SPL
 
 | |
4GW3
 
 | | Crystal Structure of the Lipase from Proteus mirabilis | | 分子名称: | CALCIUM ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Korman, T.P. | | 登録日 | 2012-08-31 | | 公開日 | 2013-02-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Proteus mirabilis Lipase, a Novel Lipase from the Proteus/Psychrophilic Subfamily of Lipase Family I.1.
Plos One, 7, 2012
|
|
4HAR
 
 | |
4GX3
 
 | |
4HBE
 
 | |
3DGN
 
 | | A non-biological ATP binding protein crystallized in the presence of 100 mM ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Simmons, C.R, Allen, J.P, Chaput, J.C. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
3DID
 
 | | Crystal structure of the F87M/L110M mutant of human transthyretin at pH 4.6 soaked | | 分子名称: | ACETATE ION, GLYCEROL, Transthyretin, ... | | 著者 | Palmieri, L.C, Freire, J.B.B, Foguel, D, Lima, L.M.T.R. | | 登録日 | 2008-06-20 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Novel Zn2+-binding sites in human transthyretin: implications for amyloidogenesis and retinol-binding protein recognition.
J.Biol.Chem., 285, 2010
|
|
4H9V
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101G/R230C with Zn2+ | | 分子名称: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | 著者 | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | 登録日 | 2012-09-25 | | 公開日 | 2012-11-07 | | 最終更新日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.971 Å) | | 主引用文献 | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4GWA
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | 分子名称: | GH7 family protein, MAGNESIUM ION | | 著者 | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | 登録日 | 2012-09-01 | | 公開日 | 2013-06-12 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HBY
 
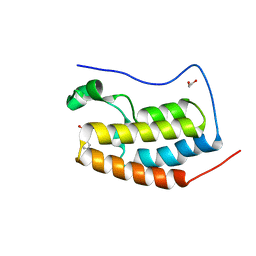 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2012-09-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4GWW
 
 | |
4GX6
 
 | |
