6UCN
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to Equilenin at 250 K | | 分子名称: | CHLORIDE ION, EQUILENIN, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-16 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MXR
 
 | |
4XR5
 
 | | X-ray structure of the unliganded thymidine phosphorylase from Salmonella typhimurium at 2.05 A resolution | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | 著者 | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | 登録日 | 2015-01-20 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | 分子名称: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | 著者 | Shi, R, Picard, M.-E, Manenda, M. | | 登録日 | 2018-11-01 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6Q2F
 
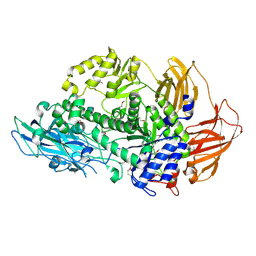 | | Structure of Rhamnosidase from Novosphingobium sp. PP1Y | | 分子名称: | Glycoside hydrolase family protein, SODIUM ION | | 著者 | Terry, B, Ha, J, Izzo, V, Sazinsky, M.H. | | 登録日 | 2019-08-07 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.20000076 Å) | | 主引用文献 | The crystal structure and insight into the substrate specificity of the alpha-L rhamnosidase RHA-P from Novosphingobium sp. PP1Y.
Arch.Biochem.Biophys., 679, 2019
|
|
6V7M
 
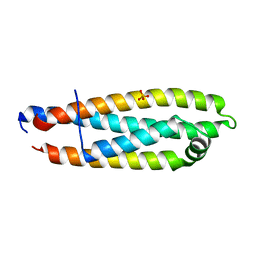 | |
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | 分子名称: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2015-02-24 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6NMV
 
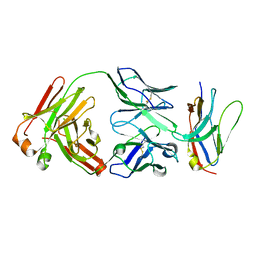 | | Non-Blocking Fab 218 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 218 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 218 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
4YEN
 
 | | Room temperature X-ray diffraction studies of cisplatin binding to HEWL in DMSO media after 14 months of crystal storage - new refinement | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | 著者 | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2015-02-24 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6NMR
 
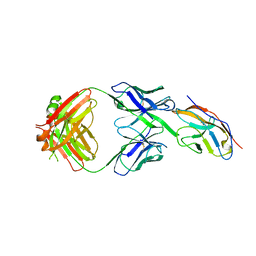 | | Blocking Fab 119 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 119 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 119 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMT
 
 | | Non-Blocking Fab 3 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 3 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 3 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
4YEO
 
 | | Triclinic HEWL co-crystallised with cisplatin, studied at a data collection temperature of 150K - new refinement | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Cisplatin, ... | | 著者 | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2015-02-24 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6NMU
 
 | | Kick-Off Fab 115 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 115 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 115 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMS
 
 | | Blocking Fab 136 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | 分子名称: | Fab 136 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 136 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | 著者 | Wibowo, A.S, Carter, J.J, Sim, J. | | 登録日 | 2019-01-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6TV2
 
 | | Heme d1 biosynthesis associated Protein NirF | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | 著者 | Kluenemann, T, Layer, G, Blankenfeldt, W. | | 登録日 | 2020-01-08 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.561 Å) | | 主引用文献 | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
6MXS
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98F,HC-G99M] | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | 著者 | Shi, R, Picard, M.-E, Manenda, M.S. | | 登録日 | 2018-10-31 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6UCY
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to 4-Androstenedione at 250 K | | 分子名称: | 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | 登録日 | 2019-09-18 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MY5
 
 | |
4YEK
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | 登録日 | 2015-02-24 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YDX
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | 分子名称: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | 著者 | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | 登録日 | 2015-02-23 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | 分子名称: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | 著者 | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2015-02-23 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6NXV
 
 | | Crystal structure of the theta class glutathione S-transferase from the citrus canker pathogen Xanthomonas axonopodis pv. citri, apo form | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Glutathione S-transferase | | 著者 | Hilario, E, De Keyser, S, Fan, L. | | 登録日 | 2019-02-10 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural and biochemical characterization of a glutathione transferase from the citrus canker pathogen Xanthomonas.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4EVI
 
 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol 9-Methyl Ether and S -Adenosyl-L-Homocysteine | | 分子名称: | 2-methoxy-4-[(1E)-3-methoxyprop-1-en-1-yl]phenol, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, ... | | 著者 | Wolters, S, Heine, A, Petersen, M. | | 登録日 | 2012-04-26 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.015 Å) | | 主引用文献 | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6OEI
 
 | | Yeast Spc42 N-terminal coiled-coil fused to PDB: 3K2N | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Spindle pole body component SPC42,Sigma-54-dependent transcriptional regulator | | 著者 | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | 登録日 | 2019-03-27 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6NV6
 
 | |
