3C6A
 
 | |
3C6Z
 
 | | HNL from Hevea brasiliensis to atomic resolution | | 分子名称: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, Hydroxynitrilase, ... | | 著者 | Schmidt, A. | | 登録日 | 2008-02-06 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Atomic resolution crystal structures and quantum chemistry meet to reveal subtleties of hydroxynitrile lyase catalysis
J.Biol.Chem., 283, 2008
|
|
3C8Q
 
 | |
3CDT
 
 | |
3CDV
 
 | |
3C80
 
 | | T4 Lysozyme mutant R96Y at room temperature | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Mooers, B.H.M. | | 登録日 | 2008-02-08 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3CDR
 
 | |
3CBL
 
 | | Crystal structure of human feline sarcoma viral oncogene homologue (v-FES) in complex with staurosporine and a consensus peptide | | 分子名称: | Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, Synthetic peptide | | 著者 | Filippakopoulos, P, Salah, E, Cooper, C, Picaud, S.S, Elkins, J.M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2008-02-22 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
3CDQ
 
 | |
1WM3
 
 | |
1WH7
 
 | | Solution structure of homeobox domain of Arabidopsis thaliana hypothetical protein F22K18.140 | | 分子名称: | ZF-HD homeobox family protein | | 著者 | Kaneno, D, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of homeobox domain of Arabidopsis thaliana hypothetical protein F22K18.140
To be Published
|
|
1WPP
 
 | | Structure of Streptococcus gordonii inorganic pyrophosphatase | | 分子名称: | CHLORIDE ION, Probable manganese-dependent inorganic pyrophosphatase, SULFATE ION, ... | | 著者 | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | 登録日 | 2004-09-09 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
1WJ1
 
 | | Solution structure of phosphotyrosine interaction domain of mouse Numb protein | | 分子名称: | Numb protein | | 著者 | Sato, M, Tomizawa, T, Koshiba, S, Tochio, N, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of phosphotyrosine interaction domain of mouse Numb protein
To be Published
|
|
1WLO
 
 | |
1IOI
 
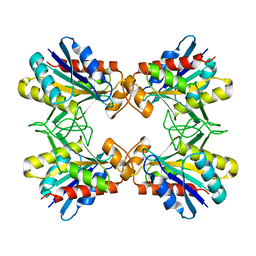 | | x-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, pyrococcus furiosus, and its cys-free mutant | | 分子名称: | PYRROLIDONE CARBOXYL PEPTIDASE | | 著者 | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | 登録日 | 2001-03-09 | | 公開日 | 2001-03-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
1WTJ
 
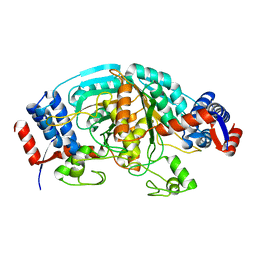 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | 分子名称: | ureidoglycolate dehydrogenase | | 著者 | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | 登録日 | 2004-11-24 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1IOT
 
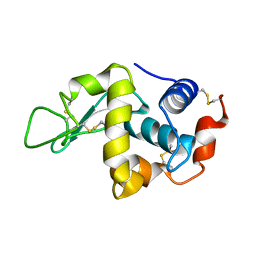 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | 分子名称: | LYSOZYME C | | 著者 | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | 登録日 | 2001-03-28 | | 公開日 | 2001-04-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
1WWN
 
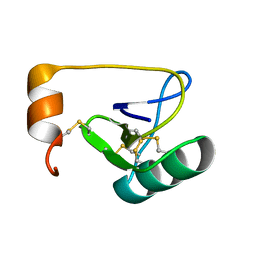 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | 分子名称: | Excitatory insect selective toxin 1 | | 著者 | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | 登録日 | 2005-01-10 | | 公開日 | 2006-01-17 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1WE6
 
 | | Solution structure of Ubiquitin-like domain in splicing factor AAL91182 | | 分子名称: | splicing factor, putative | | 著者 | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-24 | | 公開日 | 2004-11-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Ubiquitin-like domain in splicing factor AAL91182
To be Published
|
|
1WY7
 
 | |
1IXT
 
 | | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif | | 分子名称: | spasmodic protein tx9a-like protein | | 著者 | Miles, L.A, Dy, C.Y, Nielsen, J, Barnham, K.J, Hinds, M.G, Olivera, B.M, Bulaj, G, Norton, R.S. | | 登録日 | 2002-07-04 | | 公開日 | 2003-01-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif
J.Biol.Chem., 277, 2002
|
|
1WHV
 
 | | Solution structure of the RNA binding domain from hypothetical protein BAB23382 | | 分子名称: | poly(A)-specific ribonuclease | | 著者 | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RNA binding domain from hypothetical protein BAB23382
To be Published
|
|
1WIA
 
 | | Solution structure of mouse hypothetical ubiquitin-like protein BAB25500 | | 分子名称: | hypothetical ubiquitin-like protein (RIKEN cDNA 2010008E23) | | 著者 | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Hayashi, F, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of mouse hypothetical ubiquitin-like protein BAB25500
To be Published
|
|
1J2T
 
 | | Creatininase Mn | | 分子名称: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-01-11 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1X4C
 
 | | Solution structure of RRM domain in splicing factor 2 | | 分子名称: | Splicing factor, arginine/serine-rich 1 | | 著者 | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-14 | | 公開日 | 2005-11-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of RRM domain in splicing factor 2
To be Published
|
|
