5TJO
 
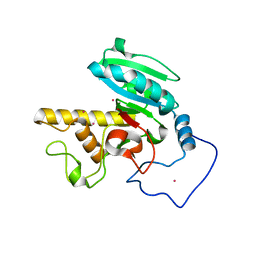 | | Crystal structure of GTB + B trisaccharide (mercury derivative) | | 分子名称: | Histo-blood group ABO system transferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | 著者 | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | 登録日 | 2016-10-04 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
5TJN
 
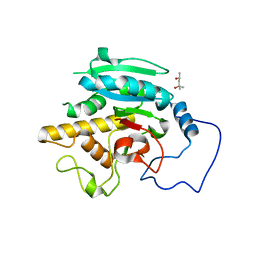 | | Crystal structure of GTB + B trisaccharide (native) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | 著者 | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | 登録日 | 2016-10-04 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
7VQI
 
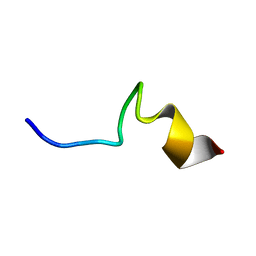 | |
5UI6
 
 | | Solution NMR Structure of Lasso Peptide Acinetodin | | 分子名称: | Acinetodin | | 著者 | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | 登録日 | 2017-01-12 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
5UI7
 
 | | Solution NMR Structure of Lasso Peptide Klebsidin | | 分子名称: | Klebsidin | | 著者 | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | 登録日 | 2017-01-13 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
1F40
 
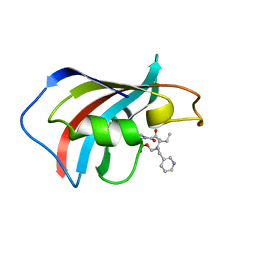 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | 分子名称: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | 著者 | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | 登録日 | 2000-06-07 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1DEF
 
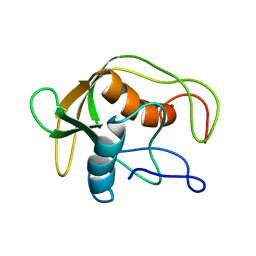 | |
5J4E
 
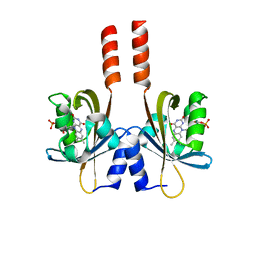 | |
5J3W
 
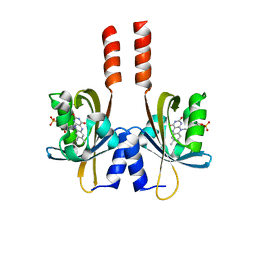 | |
3NKK
 
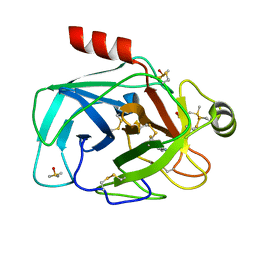 | | Trypsin in complex with fluorine containing fragment | | 分子名称: | 3-fluoro-4-methylbenzenecarboximidamide, CALCIUM ION, Cationic trypsin, ... | | 著者 | Schiering, N, Vulpetti, A, Dalvit, C. | | 登録日 | 2010-06-20 | | 公開日 | 2010-11-10 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Combined use of computational chemistry, NMR screening, and X-ray crystallography for identification and characterization of fluorophilic protein environments.
Proteins, 78, 2010
|
|
3NK8
 
 | | Trypsin in complex with fluorine-containing fragment | | 分子名称: | 4-(trifluoromethyl)-1,5,6,7-tetrahydro-2H-cyclopenta[b]pyridin-2-one, CALCIUM ION, Cationic trypsin, ... | | 著者 | Schiering, N, Vulpetti, A, Dalvit, C. | | 登録日 | 2010-06-18 | | 公開日 | 2010-11-10 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Combined use of computational chemistry, NMR screening, and X-ray crystallography for identification and characterization of fluorophilic protein environments.
Proteins, 78, 2010
|
|
5IE8
 
 | |
5JS8
 
 | | Structural Model of a Protein alpha subunit in complex with GDP obtained with SAXS and NMR residual couplings | | 分子名称: | Guanine nucleotide-binding protein G(i) subunit alpha-1 | | 著者 | Goricanec, D, Stehle, R, Grigoriu, S, Wagner, G, Hagn, F. | | 登録日 | 2016-05-07 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JR0
 
 | |
1G1N
 
 | | NICKED DECAMER DNA WITH PEG6 TETHER, NMR, 30 STRUCTURES | | 分子名称: | 5'-D(*GP*TP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*AP*CP*AP*AP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*TP*T)-3', ... | | 著者 | Bocian, W, Kozerski, L, Mazurek, A.P, Kawecki, R. | | 登録日 | 2000-10-13 | | 公開日 | 2001-03-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A nicked duplex decamer DNA with a PEG(6) tether.
Nucleic Acids Res., 29, 2001
|
|
1CWX
 
 | | SOLUTION STRUCTURE OF THE HEPATITIS C VIRUS N-TERMINAL CAPSID PROTEIN 2-45 [C-HCV(2-45)] | | 分子名称: | HEPATITIS C VIRUS CAPSID PROTEIN | | 著者 | Ladaviere, L, Deleage, G, Montserret, R, Dalbon, P, Jolivet, M, Penin, F. | | 登録日 | 1999-08-27 | | 公開日 | 1999-08-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of the Immunodominant Antigenic Region of the Hepatitis C Virus Capsid Protein by NMR
To be Published
|
|
3P8O
 
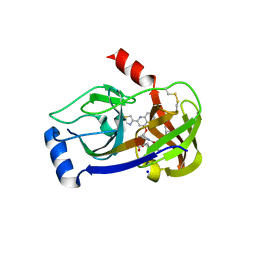 | | Crystal structure of HCV NS3/NS4A protease complexed with des-bromine analogue of BI 201335 | | 分子名称: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-4-[(7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-L-prolinamide, ... | | 著者 | Lemke, C.T. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-26 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
3P8N
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with BI 201335 | | 分子名称: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-4-[(8-bromo-7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-L-prolinamide, ... | | 著者 | Lemke, C.T. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-26 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
1F78
 
 | |
1F79
 
 | |
1F7H
 
 | |
1F7F
 
 | |
1F7I
 
 | |
1HP8
 
 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | Cx9C motif-containing protein 4 | | 著者 | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | 登録日 | 1997-08-26 | | 公開日 | 1998-03-04 | | 最終更新日 | 2019-08-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
6WPV
 
 | |
