8PBO
 
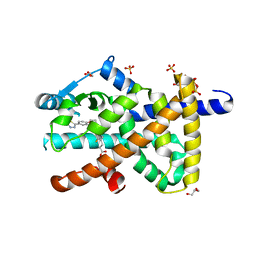 | | Deep interactome learning for generative drug design | | 分子名称: | 3-[2-fluoranyl-4-[3-[2-fluoranyl-4-(5-methyl-1,3,4-thiadiazol-2-yl)phenoxy]propoxy]phenyl]propanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | 著者 | Hakansson, M, Focht, D, Atz, K, Schneider, G. | | 登録日 | 2023-06-09 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Prospective de novo drug design with deep interactome learning.
Nat Commun, 15, 2024
|
|
7XD9
 
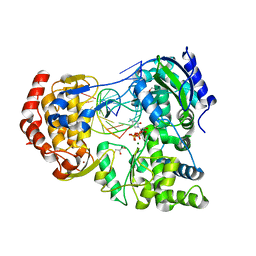 | | Crystal Structure of Dengue Virus serotype 2 (DENV2) Polymerase Elongation Complex (CTP Form) | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Wu, J, Wang, X, Gong, P. | | 登録日 | 2022-03-26 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XD8
 
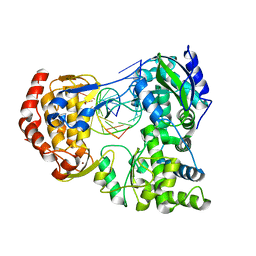 | | Crystal Structure of Dengue Virus Serotype 2 (DENV2) Polymerase Elongation Complex (Native Form) | | 分子名称: | GLYCEROL, NS5, RNA (30-mer), ... | | 著者 | Wu, J, Wang, X, Gong, P. | | 登録日 | 2022-03-26 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2MN4
 
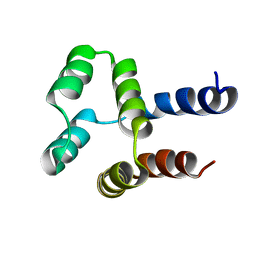 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | 分子名称: | Computational designed protein based on structure template 1cy5 | | 著者 | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | 登録日 | 2014-03-28 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
7BQE
 
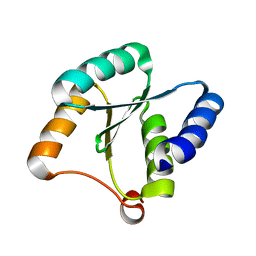 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | 分子名称: | NF3 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | 分子名称: | NF6 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
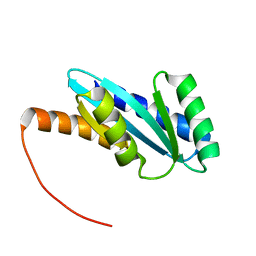 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | 分子名称: | NF4 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
4ZV6
 
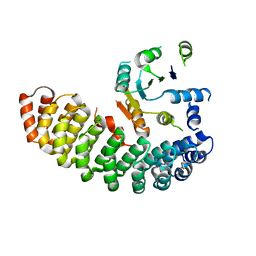 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | 分子名称: | AlphaRep-7, Octarellin V.1 | | 著者 | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | 登録日 | 2015-05-18 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6DHY
 
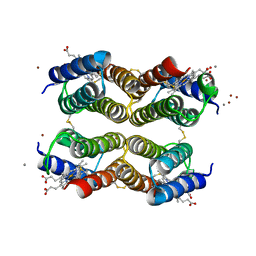 | |
6DHZ
 
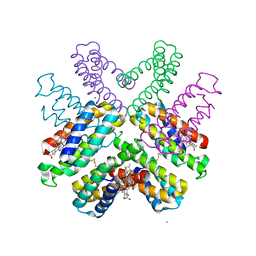 | |
6I37
 
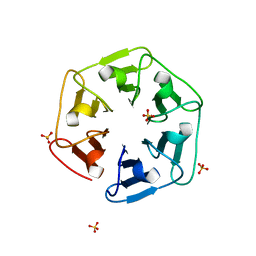 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | SULFATE ION, nv1Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I39
 
 | | Crystal structure of v31Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | MAGNESIUM ION, v31Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I38
 
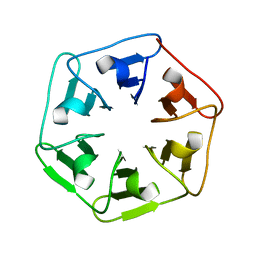 | |
6I3B
 
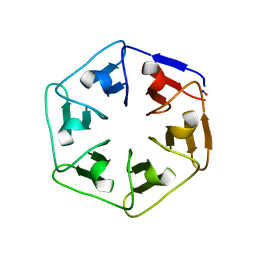 | |
6I3A
 
 | | Crystal structure of v22Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | BROMIDE ION, v22Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
7RDH
 
 | |
7BIG
 
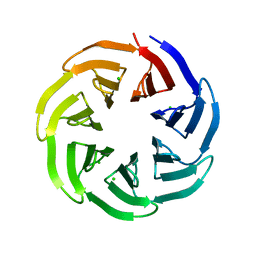 | | Crystal structure of v13WRAP-T, a 7-bladed designer protein | | 分子名称: | CHLORIDE ION, v13WRAP-T | | 著者 | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | 登録日 | 2021-01-12 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BID
 
 | | Crystal structure of v31WRAP-T, a 7-bladed designer protein | | 分子名称: | v31WRAP-T | | 著者 | Laier, I, Mylemans, B, Lee, X.Y, Voet, A.R.D. | | 登録日 | 2021-01-12 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIF
 
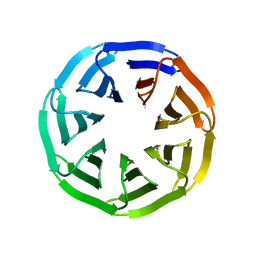 | | Crystal structure of v22WRAP-T, a 7-bladed designer protein | | 分子名称: | v22WRAP-T | | 著者 | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | 登録日 | 2021-01-12 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
5KB2
 
 | | Crystal Structure of a Tris-thiolate Zn(II)S3O Complex in a de Novo Three-stranded Coiled Coil Peptide | | 分子名称: | ZINC ION, Zn(II)(H2O)(GRAND Coil Ser-L12AL16C)3- | | 著者 | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2016-06-02 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5KB0
 
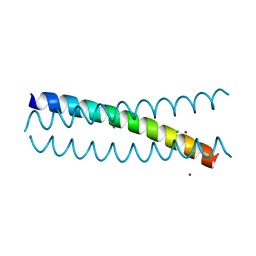 | | Crystal Structure of a Tris-thiolate Pb(II) Complex in a de Novo Three-stranded Coiled Coil Peptide | | 分子名称: | CHLORIDE ION, LEAD (II) ION, Pb(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, ... | | 著者 | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2016-06-02 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5KB1
 
 | | Crystal Structure of a Tris-thiolate Hg(II) Complex in a de Novo Three Stranded Coiled Coil Peptide | | 分子名称: | CHLORIDE ION, Hg(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, MERCURY (II) ION, ... | | 著者 | Ruckcthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2016-06-02 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5K92
 
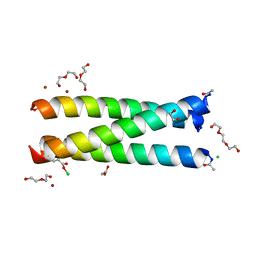 | | Crystal Structure of an apo Tris-thiolate Binding Site in a de novo Three Stranded Coiled Coil Peptide | | 分子名称: | Apo-(CSL16C)3, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2016-05-31 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
7BPN
 
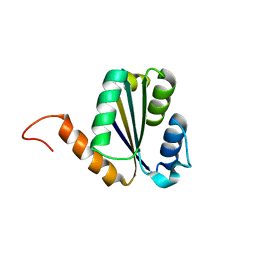 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | 分子名称: | NF7 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPP
 
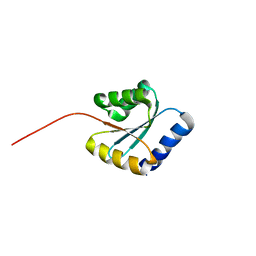 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | 分子名称: | NF5 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
