3UFA
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | 分子名称: | CHLORIDE ION, N-(3-carboxypropanoyl)-L-valyl-N-[(1S)-2-phenyl-1-phosphonoethyl]-L-prolinamide, Serine protease splA | | 著者 | Zdzalik, M, Pietrusewicz, E, Pustelny, K, Stec-Niemczyk, J, Popowicz, G.M, Potempa, J, Oleksyszyn, J, Dubin, G. | | 登録日 | 2011-10-31 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
3V3M
 
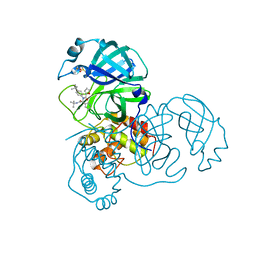 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | 著者 | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | 登録日 | 2011-12-13 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
3TEW
 
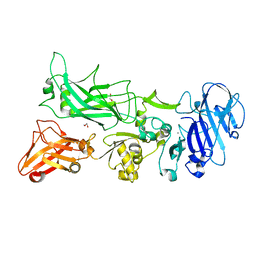 | |
3TEZ
 
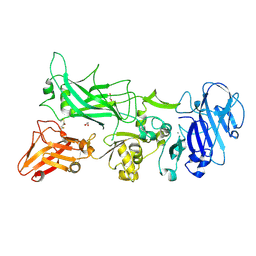 | |
1UOO
 
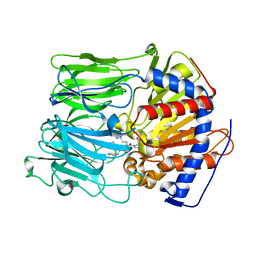 | |
1UOP
 
 | |
3TEY
 
 | |
1UOQ
 
 | |
2ZNH
 
 | | Crystal Structure of a Domain-Swapped Serpin Dimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | 著者 | Yamasaki, M, Huntington, J.A. | | 登録日 | 2008-04-25 | | 公開日 | 2008-10-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization
Nature, 455, 2008
|
|
2ASU
 
 | |
1AMO
 
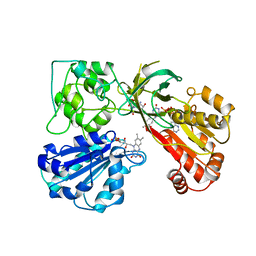 | | THREE-DIMENSIONAL STRUCTURE OF NADPH-CYTOCHROME P450 REDUCTASE: PROTOTYPE FOR FMN-AND FAD-CONTAINING ENZYMES | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Wang, M, Roberts, D.L, Paschke, R, Shea, T.M, Masters, B.S.S, Kim, J.J.P. | | 登録日 | 1997-06-17 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1LQ8
 
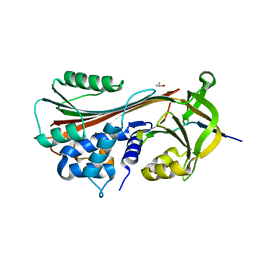 | | Crystal structure of cleaved protein C inhibitor | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Huntington, J.A, Kjellberg, M, Stenflo, J. | | 登録日 | 2002-05-09 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Structure, 11, 2003
|
|
1M8C
 
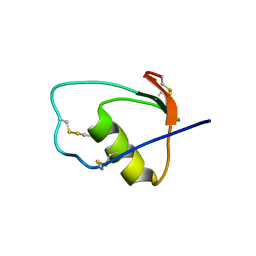 | | SOLUTION STRUCTURE OF THE T State OF TURKEY OVOMUCOID AT PH 2.5 | | 分子名称: | Ovomucoid | | 著者 | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | 登録日 | 2002-07-24 | | 公開日 | 2002-09-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1ACC
 
 | |
1AT3
 
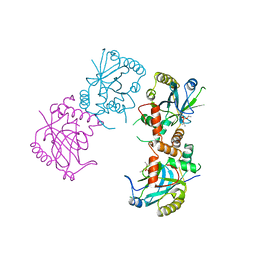 | | HERPES SIMPLEX VIRUS TYPE II PROTEASE | | 分子名称: | DIISOPROPYL PHOSPHONATE, HERPES SIMPLEX VIRUS TYPE II PROTEASE | | 著者 | Hoog, S, Smith, W.W, Qiu, X, Abdel-Meguid, S.S. | | 登録日 | 1997-08-16 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex.
Biochemistry, 36, 1997
|
|
1M8B
 
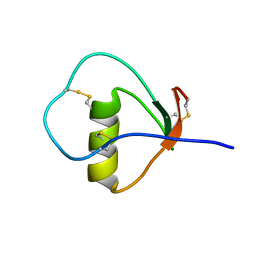 | | Solution structure of the C State of turkey ovomucoid at pH 2.5 | | 分子名称: | Ovomucoid | | 著者 | Song, J, Laskowski Jr, M, Qasim, M.A, Markley, J.L. | | 登録日 | 2002-07-24 | | 公開日 | 2002-09-04 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two conformational states of Turkey ovomucoid third domain at low pH: three-dimensional structures, internal dynamics, and interconversion kinetics and thermodynamics.
Biochemistry, 42, 2003
|
|
1MTP
 
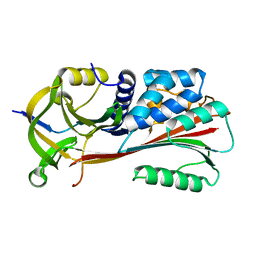 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | 分子名称: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | 著者 | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | 登録日 | 2002-09-21 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
1CMV
 
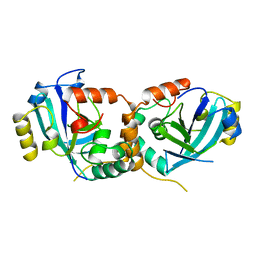 | | HUMAN CYTOMEGALOVIRUS PROTEASE | | 分子名称: | HUMAN CYTOMEGALOVIRUS PROTEASE | | 著者 | Shieh, H.-S, Kurumbail, R.G, Stevens, A.M, Stegeman, R.A, Sturman, E.J, Pak, J.Y, Wittwer, A.J, Palmier, M.O, Wiegand, R.C, Holwerda, B.C, Stallings, W.C. | | 登録日 | 1996-08-26 | | 公開日 | 1997-09-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Three-dimensional structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
1GNS
 
 | | SUBTILISIN BPN' | | 分子名称: | ACETONE, SUBTILISIN BPN' | | 著者 | Almog, O, Gallagher, D.T, Ladner, J.E, Strausberg, S, Alexander, P. | | 登録日 | 2001-10-06 | | 公開日 | 2002-06-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of Thermostability. Analysis of Stabilizing Mutations in Subtilisin Bpn'.
J.Biol.Chem., 277, 2002
|
|
1GNV
 
 | |
1R0R
 
 | | 1.1 Angstrom Resolution Structure of the Complex Between the Protein Inhibitor, OMTKY3, and the Serine Protease, Subtilisin Carlsberg | | 分子名称: | CALCIUM ION, Ovomucoid, subtilisin carlsberg | | 著者 | Horn, J.R, Ramaswamy, S, Murphy, K.P. | | 登録日 | 2003-09-22 | | 公開日 | 2003-11-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structure and energetics of protein-protein interactions: the role of conformational heterogeneity in OMTKY3 binding to serine proteases
J.Mol.Biol., 331, 2003
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | 分子名称: | OVOMUCOID (THIRD DOMAIN) | | 著者 | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | 登録日 | 1995-10-11 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1OMT
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | 分子名称: | OVOMUCOID (THIRD DOMAIN) | | 著者 | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | 登録日 | 1995-10-11 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1RRE
 
 | | Crystal structure of E.coli Lon proteolytic domain | | 分子名称: | ATP-dependent protease La, SULFATE ION | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | 登録日 | 2003-12-08 | | 公開日 | 2004-02-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | 分子名称: | ATP-dependent protease La, SULFATE ION | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | 登録日 | 2003-12-08 | | 公開日 | 2003-12-23 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
