4ZBR
 
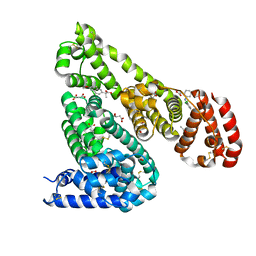 | | Crystal Structure of Equine Serum Albumin in complex with Diclofenac and Naproxen | | 分子名称: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | 著者 | Sekula, B, Bujacz, A, Bujacz, G. | | 登録日 | 2015-04-15 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural Insights into the Competitive Binding of Diclofenac and Naproxen by Equine Serum Albumin.
J.Med.Chem., 59, 2016
|
|
3VOD
 
 | |
3VTV
 
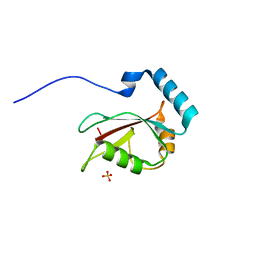 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | 分子名称: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | 著者 | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | 登録日 | 2012-06-08 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
5LUL
 
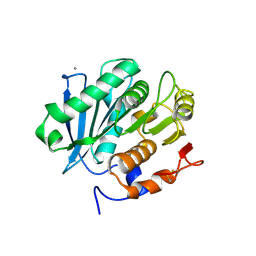 | | Structure of a triple variant of cutinase 2 from Thermobifida cellulosilytica | | 分子名称: | CALCIUM ION, CHLORIDE ION, Cutinase 2 | | 著者 | Hromic, A, Lyskowski, A, Gruber, K. | | 登録日 | 2016-09-09 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
5AAO
 
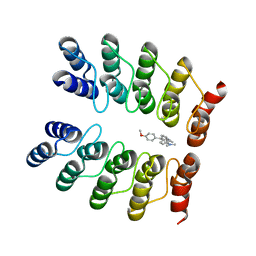 | | Crystal structure of fluorogen-activating designed ankyrin repeat protein (DARPin) dimer in complex with malachite green | | 分子名称: | 4-[[4-(dimethylamino)cyclohexa-2,5-dien-1-ylidene]-(4-methoxyphenyl)methyl]-N,N-dimethyl-aniline, FAD3210 | | 著者 | Batyuk, A, Schuetz, M, Kummer, L, Wu, Y, Mittl, P, Plueckthun, A. | | 登録日 | 2015-07-27 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Generation of Fluorogen-Activating Designed Ankyrin Repeat Proteins (FADAs) as Versatile Sensor Tools.
J. Mol. Biol., 428, 2016
|
|
8PIU
 
 | |
5LPU
 
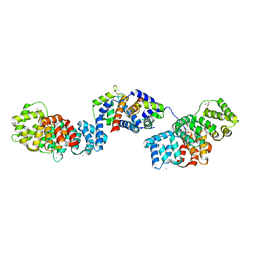 | | Crystal structure of Annexin A2 complexed with S100A4 | | 分子名称: | Annexin A2, CALCIUM ION, GLYCEROL, ... | | 著者 | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | 登録日 | 2016-08-15 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5AK7
 
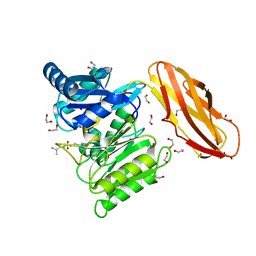 | | Structure of wt Porphyromonas gingivalis peptidylarginine deiminase | | 分子名称: | 1,2-ETHANEDIOL, ALANINE, ARGININE, ... | | 著者 | Kopec, J, Montgomery, A, Shrestha, L, Kiyani, W, Nowak, R, Burgess-Brown, N, Venables, P.J, Yue, W.W. | | 登録日 | 2015-03-02 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal Structure of Porphyromonas Gingivalis Peptidylarginine Deiminase: Implications for Autoimmunity in Rheumatoid Arthritis.
Ann.Rheum.Dis., 75, 2016
|
|
3W6H
 
 | | Crystal structure of 19F probe-labeled hCAI in complex with acetazolamide | | 分子名称: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 1, ... | | 著者 | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | 登録日 | 2013-02-14 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.964 Å) | | 主引用文献 | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
4R0I
 
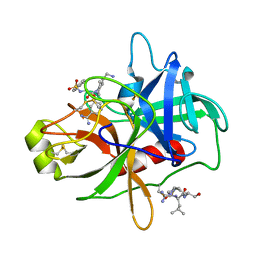 | | CRYSTAL STRUCTURE of MATRIPTASE in COMPLEX WITH INHIBITOR | | 分子名称: | 3-({(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-[(naphthalen-2-ylsulfonyl)amino]-3-oxopropyl}oxy)benzenecarboximidamide, SERINE PROTEASE, MATRIPTASE, ... | | 著者 | Rao, K.N, Ashok, K.N, Chakshusmathi, G, Rajeev, G, Subramanya, H. | | 登録日 | 2014-07-31 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5LXL
 
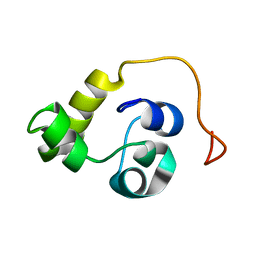 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | 分子名称: | Decoration protein | | 著者 | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | 登録日 | 2016-09-22 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
6MAZ
 
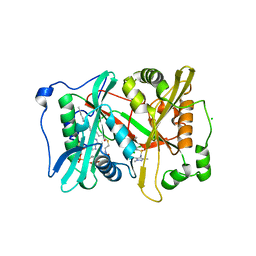 | |
4R6Q
 
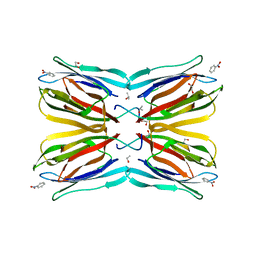 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | 分子名称: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | 著者 | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | 登録日 | 2014-08-26 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QR3
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
5A8Z
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(4R)-7-methyl-2,5-bis(oxidanylidene)-1-[3-(trifluoromethyl)phenyl]-3,4,6,8-tetrahydropyrimido[4,5-d]pyridazin-4-yl]benzenecarbonitrile, Neutrophil elastase, ... | | 著者 | von Nussbaum, F, Li, V.M, Meibom, D, Anlauf, S, Bechem, M, Delbeck, M, Gerisch, M, Harrenga, A, Karthaus, D, Lang, D, Lustig, K, Mittendorf, J, Schaefer, M, Schaefer, S, Schamberger, J. | | 登録日 | 2015-07-17 | | 公開日 | 2016-08-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Potent and Selective Human Neutrophil Elastase Inhibitors with Novel Equatorial Ring Topology: in vivo Efficacy of the Polar Pyrimidopyridazine BAY-8040 in a Pulmonary Arterial Hypertension Rat Model.
ChemMedChem, 11, 2016
|
|
3WV3
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-2-carboxamide | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | 著者 | Oki, H, Tanaka, Y. | | 登録日 | 2014-05-12 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
5BY3
 
 | | A novel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans | | 分子名称: | BtGH115A, NICKEL (II) ION, SULFATE ION | | 著者 | Lammerts van Bueren, A, Davies, G.J, Turkenburg, J.P. | | 登録日 | 2015-06-10 | | 公開日 | 2015-07-22 | | 最終更新日 | 2015-12-09 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural and Functional Characterization of a Novel Family GH115 4-O-Methyl-alpha-Glucuronidase with Specificity for Decorated Arabinogalactans.
J.Mol.Biol., 427, 2015
|
|
5MRS
 
 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | 分子名称: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF
To Be Published
|
|
3W6I
 
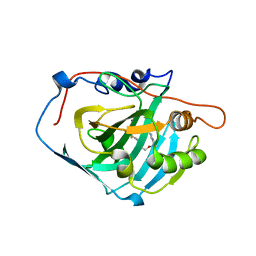 | | Crystal structure of 19F probe-labeled hCAI | | 分子名称: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | 著者 | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | 登録日 | 2013-02-14 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
4RCL
 
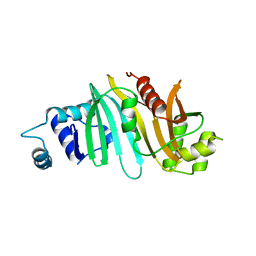 | |
3W7A
 
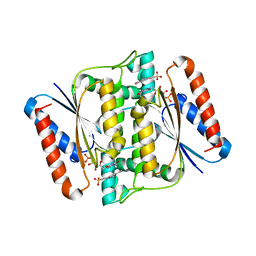 | | Crystal Structure of azoreductase AzrC fin complex with sulfone-modified azo dye Acid Red 88 | | 分子名称: | 4-[(E)-(2-hydroxynaphthalen-1-yl)diazenyl]naphthalene-1-sulfonic acid, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Yu, J, Ogata, D, Ooi, T, Yao, M. | | 登録日 | 2013-02-27 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5MTR
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT512 | | 分子名称: | 2-[4-[(4-cyclopentyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | 著者 | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | 登録日 | 2017-01-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
6LWA
 
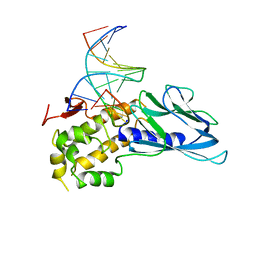 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2020-02-07 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWF
 
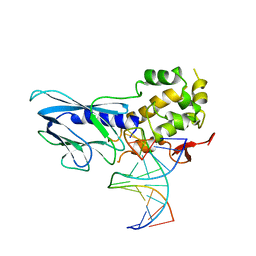 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | 著者 | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2020-02-07 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
