3NMU
 
 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | 分子名称: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | 著者 | Li, H, Xue, S, Wang, R. | | 登録日 | 2010-06-22 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.729 Å) | | 主引用文献 | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
6HIU
 
 | | Cytochrome P460 from Methylococcus capsulatus (Bath) | | 分子名称: | Cytochrome P460, GLYCEROL, HEME C, ... | | 著者 | Adams, H, Chicano, T.M, Hough, M.A. | | 登録日 | 2018-08-31 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | One fold, two functions: cytochrome P460 and cytochromec'-beta from the methanotrophMethylococcus capsulatus(Bath).
Chem Sci, 10, 2019
|
|
5YY6
 
 | | Crystal structure of Arabidopsis thaliana HPPD truncated mutant complexed with Benquitrione | | 分子名称: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, W.C, Chen, J.N, Yang, G.F. | | 登録日 | 2017-12-08 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Research (Wash D C), 2019, 2019
|
|
6HQ4
 
 | |
3X39
 
 | | Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551 | | 分子名称: | Cytochrome c-551, HEME C | | 著者 | Nagao, S, Ueda, M, Osuka, H, Komori, H, Kamikubo, H, Kataoka, M, Higuchi, Y, Hirota, S. | | 登録日 | 2015-01-16 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Plos One, 10, 2015
|
|
5VAP
 
 | | Crystal structure of eVP30 C-terminus and eNP peptide | | 分子名称: | Minor nucleoprotein VP30, NP | | 著者 | XU, W, WU, C, Leung, D.W, Amarasinghe, G.K. | | 登録日 | 2017-03-27 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis.
Nat Commun, 8, 2017
|
|
5FAW
 
 | | T502A mutant of choline TMA-lyase | | 分子名称: | CHOLINE ION, Choline trimethylamine-lyase, MALONATE ION, ... | | 著者 | Funk, M.A, Drennan, C.L. | | 登録日 | 2015-12-12 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.852 Å) | | 主引用文献 | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
2W97
 
 | | Crystal Structure of eIF4E Bound to Glycerol and eIF4G1 peptide | | 分子名称: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4 GAMMA 1, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, GLYCEROL, ... | | 著者 | Brown, C.J, Verma, C.S, Walkinshaw, M.D, Lane, D.P. | | 登録日 | 2009-01-22 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site.
Cell Cycle, 8, 2009
|
|
4AEC
 
 | | Crystal Structure of the Arabidopsis thaliana O-Acetyl-Serine-(Thiol)- Lyase C | | 分子名称: | ACETATE ION, CYSTEINE SYNTHASE, MITOCHONDRIAL, ... | | 著者 | Feldman-Salit, A, Wirtz, M, Lenherr, E.D, Throm, C, Hothorn, M, Scheffzek, K, Hell, R, Wade, R.C. | | 登録日 | 2012-01-09 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Allosterically Gated Enzyme Dynamics in the Cysteine Synthase Complex Regulate Cysteine Biosynthesis in Arabidopsis Thaliana.
Structure, 20, 2012
|
|
5V6N
 
 | |
2E9T
 
 | | Foot-and-mouth disease virus RNA-polymerase RNA dependent in complex with a template-primer RNA and 5F-UTP | | 分子名称: | 5'-R(*GP*GP*GP*CP*CP*CP*(5FU))-3', 5'-R(P*UP*AP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | 著者 | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | 登録日 | 2007-01-26 | | 公開日 | 2007-06-26 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4DDI
 
 | |
7FWZ
 
 | | Crystal Structure of human FABP4 in complex with 4-[3-(trifluoromethyl)-5,6,7,8-tetrahydro-4H-cyclohepta[c]pyrazol-1-yl]butanoic acid | | 分子名称: | 4-[3-(trifluoromethyl)-5,6,7,8-tetrahydrocyclohepta[c]pyrazol-1(4H)-yl]butanoic acid, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
7FY6
 
 | | Crystal Structure of human FABP4 in complex with 6-phenyl-11H-pyrimido[4,5-c][2]benzazepin-3-amine | | 分子名称: | (10S)-10-phenyl-10,11-dihydro-5H-pyrimido[4,5-c][2]benzazepin-2-amine, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Fryer, R, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
1RAP
 
 | |
3MWU
 
 | | Activated Calcium-Dependent Protein Kinase 1 from Cryptosporidium parvum (CpCDPK1) in complex with bumped kinase inhibitor RM-1-95 | | 分子名称: | 3-(naphthalen-1-ylmethyl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, Calmodulin-domain protein kinase 1 | | 著者 | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | 登録日 | 2010-05-06 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of Potent and Selective Inhibitors of Calcium-Dependent Protein Kinase 1 (CDPK1) from C. parvum and T. gondii.
ACS Med Chem Lett, 1, 2010
|
|
5FAY
 
 | | Y208F mutant of choline TMA-lyase | | 分子名称: | CHOLINE ION, Choline trimethylamine-lyase, MALONATE ION, ... | | 著者 | Funk, M.A, Drennan, C.L. | | 登録日 | 2015-12-12 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
1PTG
 
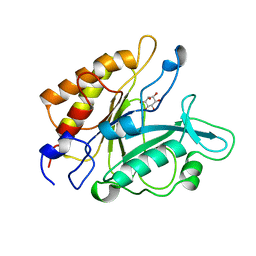 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C IN COMPLEX WITH MYO-INOSITOL | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | 著者 | Heinz, D.W, Ryan, M, Bullock, T.L, Griffith, O.H. | | 登録日 | 1995-05-24 | | 公開日 | 1996-07-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol.
EMBO J., 14, 1995
|
|
7XWN
 
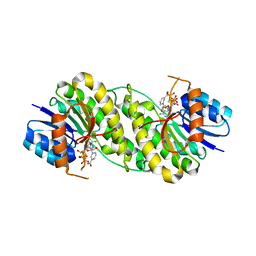 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
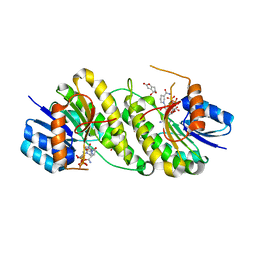 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
3NL0
 
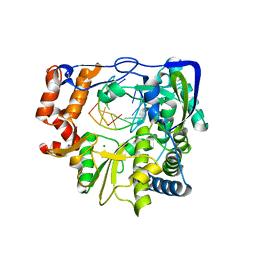 | | Mutant P44S M296I of Foot-and-mouth disease Virus RNA-dependent RNA polymerase | | 分子名称: | 3D polymerase, 5'-R(*G*GP*GP*CP*CP*C)-3', 5'-R(P*UP*GP*GP*GP*CP*CP*C)-3', ... | | 著者 | Agudo, R, Ferrer-Orta, C, Arias, A, Perez-Luque, R, Verdaguer, N, Domingo, E. | | 登録日 | 2010-06-21 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape.
Plos Pathog., 6, 2010
|
|
6CV9
 
 | |
5GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, GLUTATHIONYLSPERMIDINE COMPLEX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, GLUTATHIONYLSPERMIDINE DISULFIDE | | 著者 | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | 登録日 | 1997-02-12 | | 公開日 | 1997-08-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
6ZPP
 
 | |
1NMI
 
 | | Solution structure of the imidazole complex of iso-1 cytochrome c | | 分子名称: | Cytochrome c, iso-1, HEME C, ... | | 著者 | Yao, Y, Tong, Y, Liu, G, Wang, J, Zheng, J, Tang, W. | | 登録日 | 2003-01-10 | | 公開日 | 2003-02-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the imidazole complex of iso-1 cytochrome c
To be Published
|
|
