3D6I
 
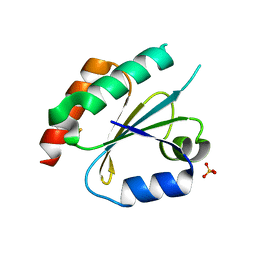 | | Structure of the Thioredoxin-like Domain of Yeast Glutaredoxin 3 | | 分子名称: | Monothiol glutaredoxin-3, SULFATE ION | | 著者 | Lebioda, L, Gibson, L.M, Dingra, N.N, Outten, C.E. | | 登録日 | 2008-05-19 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the thioredoxin-like domain of yeast glutaredoxin 3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
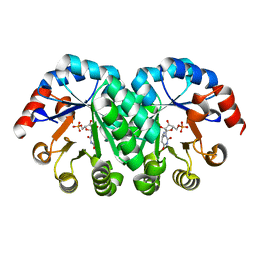
2ZZ5
 
 | |
3LL9
 
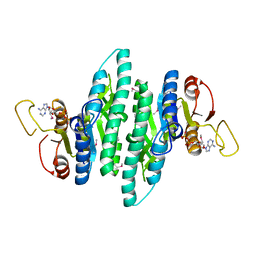 | |
4R2N
 
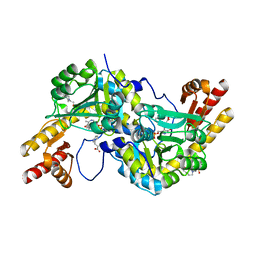 | | Crystal structure of Rv3772 in complex with its substrate | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | 登録日 | 2014-08-12 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
4TTL
 
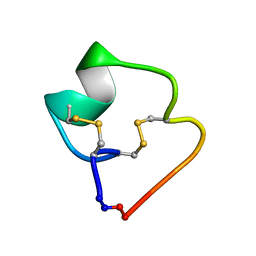 | |
6M4Z
 
 | |
4U2P
 
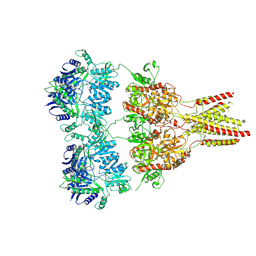 | |
7Z79
 
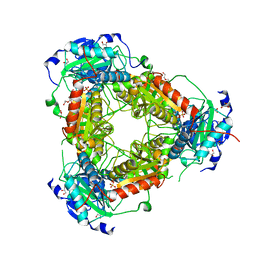 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | 分子名称: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-03-15 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
4U2Q
 
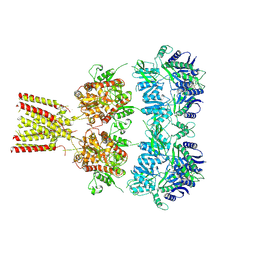 | | Full-length AMPA subtype ionotropic glutamate receptor GluA2 in complex with partial agonist kainate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | 著者 | Duerr, K.L, Chen, L, Gouaux, E. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.5247 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
6VR7
 
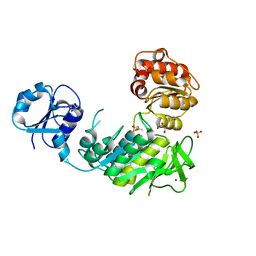 | | Structure of a pseudomurein peptide ligase type C from Methanothermus fervidus | | 分子名称: | ACETOACETIC ACID, GLYCEROL, Mur ligase middle domain protein, ... | | 著者 | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | 登録日 | 2020-02-06 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry
FEMS Microbes, 2, 2021
|
|
6VR8
 
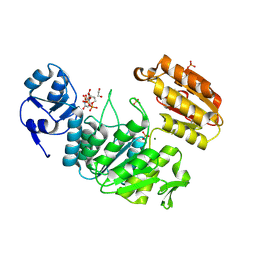 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | 著者 | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | 登録日 | 2020-02-06 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
5X6L
 
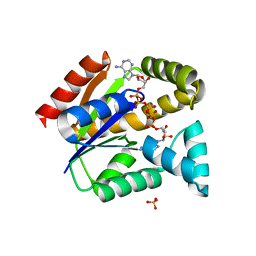 | |
5NF6
 
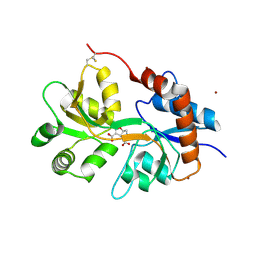 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with CIP-AS at 2.55 A resolution | | 分子名称: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | 登録日 | 2017-03-13 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NF5
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with CIP-AS at 2.85 A resolution | | 分子名称: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | 登録日 | 2017-03-13 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
7NPA
 
 | |
7UFP
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | 分子名称: | Mur ligase middle domain protein, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | 著者 | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | 登録日 | 2022-03-23 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
2YEN
 
 | | Solution structure of the skeletal muscle and neuronal voltage gated sodium channel antagonist mu-conotoxin CnIIIC | | 分子名称: | Mu-conotoxin CnIIIC | | 著者 | Favreau, P, Benoit, E, Hocking, H.G, Carlier, L, D'hoedt, D, Leipold, E, Markgraf, R, Schlumberger, S, Cordova, M.A, Gaertner, H, Paolini-Bertrand, M, Hartley, O, Tytgat, J, Heinemann, S.H, Bertrand, D, Boelens, R, Stocklin, R, Molgo, J. | | 登録日 | 2011-03-28 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Novel Mu-Conopeptide, Cniiic, Exerts Potent and Preferential Inhibition of Na(V) 1.2/1.4 Channels and Blocks Neuronal Nicotinic Acetylcholine Receptors.
Br.J.Pharmacol., 166, 2012
|
|
8ONL
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONJ
 
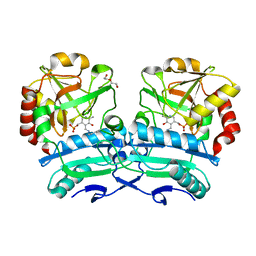 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | 分子名称: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
7TZI
 
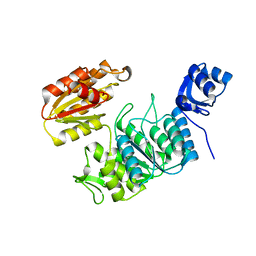 | | Structure of a pseudomurein peptide ligase type E from Methanothermobacter thermautotrophicus | | 分子名称: | Mur ligase family protein | | 著者 | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | 登録日 | 2022-02-15 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.911 Å) | | 主引用文献 | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
5IKB
 
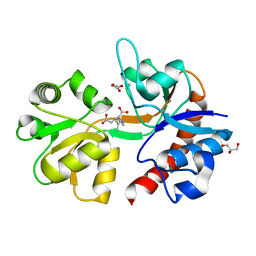 | | Crystal structure of the kainate receptor GluK4 ligand binding domain in complex with kainate | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, GLYCEROL, Glutamate receptor ionotropic, ... | | 著者 | Kristensen, O, Kristensen, L.B, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-03-03 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The Structure of a High-Affinity Kainate Receptor: GluK4 Ligand-Binding Domain Crystallized with Kainate.
Structure, 24, 2016
|
|
2OWX
 
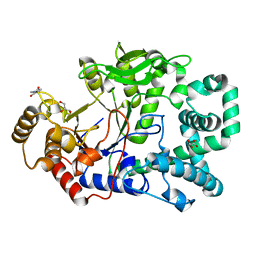 | | THERMUS THERMOPHILUS AMYLOMALTASE AT pH 5.6 | | 分子名称: | 4-alpha-glucanotransferase, GLYCEROL, MALONATE ION | | 著者 | Barends, T.R.M, Kaper, T, Bultema, J.J, Dijkhuizen, L, van der Maarel, J.E.C, Dijkstra, B.W. | | 登録日 | 2007-02-17 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase.
J.Biol.Chem., 282, 2007
|
|
7JT8
 
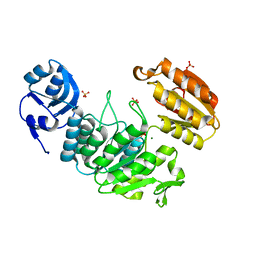 | | Apo structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | 分子名称: | MAGNESIUM ION, Mur ligase middle domain protein, SULFATE ION | | 著者 | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | 登録日 | 2020-08-17 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
6ZLF
 
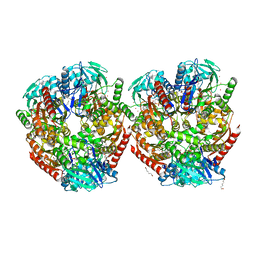 | | Aerobic crystal structure of F420H2-Oxidase from Methanothermococcus thermolithotrophicus at 1.8A resolution under 125 bars of krypton | | 分子名称: | CHLORIDE ION, Coenzyme F420H2 oxidase (FprA), FLAVIN MONONUCLEOTIDE, ... | | 著者 | Engilberge, S, Wagner, T, Carpentier, P, Girard, E, Shima, S. | | 登録日 | 2020-06-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Krypton-derivatization highlights O 2 -channeling in a four-electron reducing oxidase.
Chem.Commun.(Camb.), 56, 2020
|
|
5KCA
 
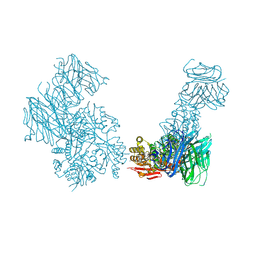 | | Crystal structure of the Cbln1 C1q domain trimer in complex with the amino-terminal domain (ATD) of iGluR Delta-2 (GluD2) | | 分子名称: | CALCIUM ION, Cerebellin-1,Cerebellin-1,Cerebellin-1,Glutamate receptor ionotropic, delta-2 | | 著者 | Elegheert, J, Aricescu, A.R. | | 登録日 | 2016-06-05 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
