3M2V
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | 分子名称: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | 登録日 | 2010-03-08 | | 公開日 | 2010-09-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | 分子名称: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | 分子名称: | Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
1EJE
 
 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | 著者 | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2000-03-02 | | 公開日 | 2000-10-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
4E0X
 
 | |
3LHZ
 
 | | Crystal structure of the mutant V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LHV
 
 | | Crystal structure of the mutant V182A.I199A.V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
1EO1
 
 | |
3LHY
 
 | | Crystal structure of the mutant I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LTP
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-02-16 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
1EG5
 
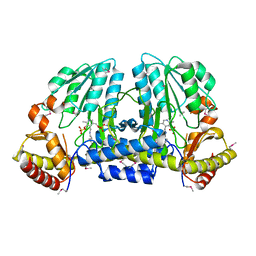 | | NIFS-LIKE PROTEIN | | 分子名称: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | 登録日 | 2000-02-13 | | 公開日 | 2000-04-02 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
3LTY
 
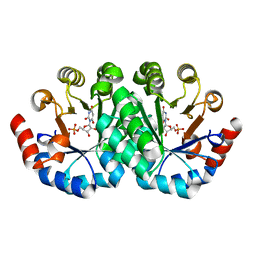 | | Crystal structure of the mutant V182A,I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-02-16 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LV5
 
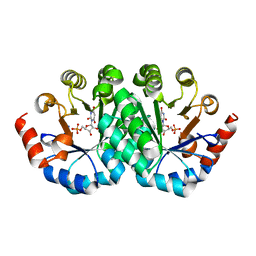 | | Crystal structure of the mutant I199E of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-02-19 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.444 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LV6
 
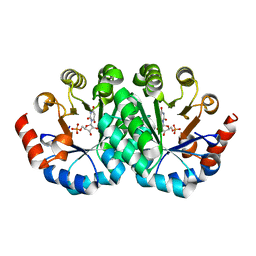 | | Crystal structure of the mutant I218F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-02-19 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3M41
 
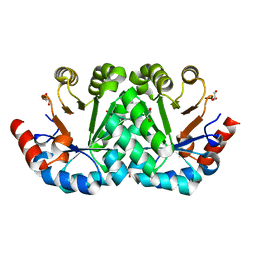 | | Crystal structure of the mutant V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | 分子名称: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-03-10 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
7Z2R
 
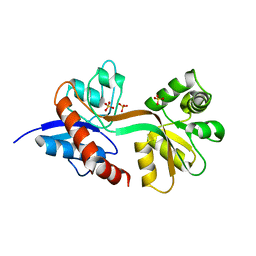 | | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics | | 分子名称: | Glutamate receptor ionotropic, delta-1, SULFATE ION | | 著者 | Masternak, M, Laulumaa, S, Kastrup, J.S. | | 登録日 | 2022-02-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.574 Å) | | 主引用文献 | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics.
Febs J., 290, 2023
|
|
7TB6
 
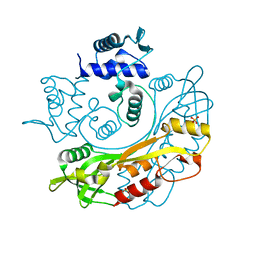 | | Structure of S. maltophilia CapW | | 分子名称: | S. maltophilia CapW, SULFATE ION | | 著者 | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | 登録日 | 2021-12-21 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
7SUC
 
 | |
1ECX
 
 | | NIFS-LIKE PROTEIN | | 分子名称: | AMINOTRANSFERASE, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Kaiser, J.T, Clausen, T.C, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | 登録日 | 2000-01-26 | | 公開日 | 2000-03-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
5Y4U
 
 | | Crystal structure of Grx domain of Grx3 from Saccharomyces cerevisiae | | 分子名称: | Monothiol glutaredoxin-3 | | 著者 | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | 登録日 | 2017-08-05 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
6LX2
 
 | | Potato D-enzyme complexed with CA26 | | 分子名称: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, 4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose, ... | | 著者 | Unno, H, Imamura, K. | | 登録日 | 2020-02-10 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
6S34
 
 | |
4YP5
 
 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | 著者 | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | 登録日 | 2015-03-12 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3G0Z
 
 | | Structure of S. pombe Pop2p - Zn2+ and Mn2+ bound form | | 分子名称: | CCR4-Not complex subunit Caf1, MANGANESE (II) ION, ZINC ION | | 著者 | Andersen, K.R, Jonstrup, A.T, Van, L.B, Brodersen, D.E. | | 登録日 | 2009-01-29 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site
Rna, 15, 2009
|
|
