8I5S
 
 | | Crystal structure of TxGH116 D593N acid/base mutant from Thermoanaerobacterium xylanolyticum with 2-deoxy-2-fluoroglucoside | | 分子名称: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | 著者 | Pengthaisong, S, Ketudat Cairns, J.R. | | 登録日 | 2023-01-26 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Reaction Mechanism of Glycoside Hydrolase Family 116 Utilizes Perpendicular Protonation.
Acs Catalysis, 13, 2023
|
|
8I5R
 
 | |
8I5Q
 
 | |
8I5P
 
 | |
8I5O
 
 | |
8I5L
 
 | |
8I5G
 
 | | Structure of human Nav1.7 in complex with PF-05089771 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-25 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5C
 
 | | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK) | | 分子名称: | Beta-2-microglobulin, MHC class I antigen (Fragment), TCR alpha chain, ... | | 著者 | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | 登録日 | 2023-01-24 | | 公開日 | 2023-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
8I5B
 
 | | Structure of human Nav1.7 in complex with bupivacaine | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5A
 
 | | N-acetyl-(R)-beta-phenylalanine acylase, 2.75 angstrom resolution | | 分子名称: | N-acetyl-(R)-beta-phenylalanine acylase | | 著者 | Kato, Y, Natsume, R. | | 登録日 | 2023-01-24 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8I59
 
 | | N-acetyl-(R)-beta-phenylalanine acylase, selenomethionyl derivative | | 分子名称: | N-acetyl-(R)-beta-phenylalanine acylase | | 著者 | Kato, Y, Natsume, R. | | 登録日 | 2023-01-24 | | 公開日 | 2023-03-15 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8I58
 
 | | Uroporphyrin I (UPI)-bound CfbA | | 分子名称: | 3-[(1Z,9Z)-3,8,13,18-tetrakis(2-hydroxy-2-oxoethyl)-7,12,17-tris(3-hydroxy-3-oxopropyl)-21,23-dihydroporphyrin-2-yl]propanoic acid, Sirohydrochlorin cobaltochelatase | | 著者 | Ogawa, S, Fujishiro, T. | | 登録日 | 2023-01-24 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Substrate selectivity of CfbA
To be published
|
|
8I57
 
 | | Uroporphyrin III (UPIII)-bound CfbA | | 分子名称: | 3,3',3'',3'''-[3,8,13,17-tetrakis(carboxymethyl)porphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Sirohydrochlorin cobaltochelatase | | 著者 | Ogawa, S, Fujishiro, T. | | 登録日 | 2023-01-24 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Substrate selectivity of CfbA
To be published
|
|
8I56
 
 | |
8I55
 
 | |
8I50
 
 | |
8I4S
 
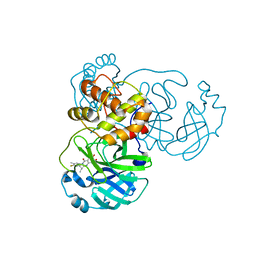 | | the complex structure of SARS-CoV-2 Mpro with D8 | | 分子名称: | 3-(4-fluoranyl-3-methyl-phenyl)-2-(2-methylpropyl)-5,6,7-tris(oxidanyl)quinazolin-4-one, ORF1a polyprotein | | 著者 | Lu, M. | | 登録日 | 2023-01-21 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of quinazolin-4-one-based non-covalent inhibitors targeting the severe acute respiratory syndrome coronavirus 2 main protease (SARS-CoV-2 M pro ).
Eur.J.Med.Chem., 257, 2023
|
|
8I4Q
 
 | |
8I4P
 
 | |
8I4K
 
 | | Structure of Azami Red1.0, a red fluorescent protein engineered from Azami Green | | 分子名称: | Azami Red1.0, CALCIUM ION | | 著者 | Otsubo, S, Takekawa, N, Imamura, H, Imada, K. | | 登録日 | 2023-01-19 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Red fluorescent proteins engineered from green fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I4J
 
 | |
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | 登録日 | 2023-01-19 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
8I48
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-01-18 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8I47
 
 | |
8I42
 
 | |
