3WFI
 
 | | The crystal structure of D-mandelate dehydrogenase | | 分子名称: | 2-dehydropantoate 2-reductase | | 著者 | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | 登録日 | 2013-07-19 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
7UV9
 
 | | KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate | | 分子名称: | DNA (185-MER), FE (III) ION, Histone H2A type 1, ... | | 著者 | Spangler, C.J, Skrajna, A, Foley, C.A, Budziszewski, G.R, Azzam, D.N, James, L.I, Frye, S.V, McGinty, R.K. | | 登録日 | 2022-04-29 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
1N0H
 
 | | Crystal Structure of Yeast Acetohydroxyacid Synthase in Complex with a Sulfonylurea Herbicide, Chlorimuron Ethyl | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, 4-{[(4'-AMINO-2'-METHYLPYRIMIDIN-5'-YL)METHYL]AMINO}PENT-3-ENYL DIPHOSPHATE, ... | | 著者 | Pang, S.S, Guddat, L.W, Duggleby, R.G. | | 登録日 | 2002-10-14 | | 公開日 | 2003-01-07 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase
J.BIOL.CHEM., 278, 2003
|
|
7DDP
 
 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | 登録日 | 2020-10-29 | | 公開日 | 2021-05-19 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7DDO
 
 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | 登録日 | 2020-10-29 | | 公開日 | 2021-05-19 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
1NZJ
 
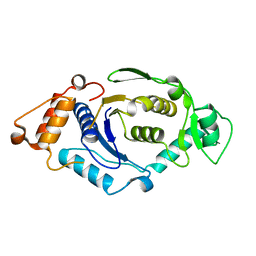 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | 分子名称: | Hypothetical protein yadB, ZINC ION | | 著者 | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | 登録日 | 2003-02-18 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
5B7B
 
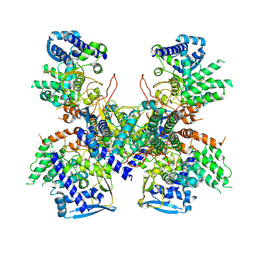 | | Crystal structure of Nucleoprotein-nucleozin complex | | 分子名称: | Nucleoprotein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl](5-methyl-3-phenyl-1,2-oxazol-4-yl)methanone | | 著者 | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | 登録日 | 2016-06-06 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Characterization of H1N1 Nucleoprotein-Nucleozin Binding Sites
Sci Rep, 6, 2016
|
|
1NNY
 
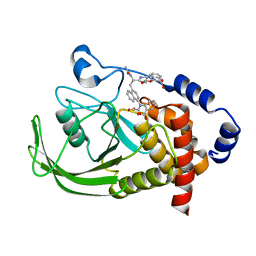 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 23 Using a Linked-Fragment Strategy | | 分子名称: | 3-({5-[(N-ACETYL-3-{4-[(CARBOXYCARBONYL)(2-CARBOXYPHENYL)AMINO]-1-NAPHTHYL}-L-ALANYL)AMINO]PENTYL}OXY)-2-NAPHTHOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | 登録日 | 2003-01-14 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1N8V
 
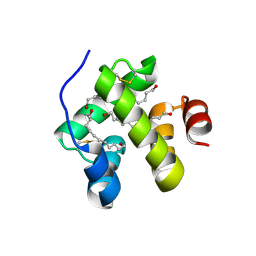 | | Chemosensory Protein in complex with bromo-dodecanol | | 分子名称: | BROMO-DODECANOL, chemosensory protein | | 著者 | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | 登録日 | 2002-11-21 | | 公開日 | 2003-04-01 | | 最終更新日 | 2017-02-01 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OZG
 
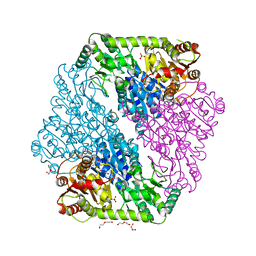 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate | | 分子名称: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | 著者 | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | 登録日 | 2003-04-09 | | 公開日 | 2003-11-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
5G48
 
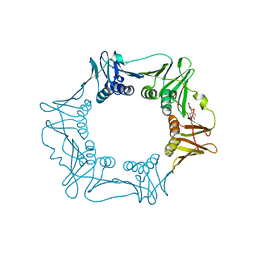 | | H.pylori Beta clamp in complex with Diflunisal | | 分子名称: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | 著者 | Pandey, P, Gourinath, S. | | 登録日 | 2016-05-06 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
1NPH
 
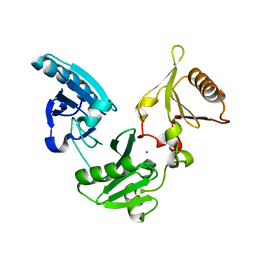 | | Gelsolin Domains 4-6 in Active, Actin Free Conformation Identifies Sites of Regulatory Calcium Ions | | 分子名称: | CALCIUM ION, Gelsolin | | 著者 | Kolappan, S, Gooch, J.T, Weeds, A.G, McLaughlin, P.J. | | 登録日 | 2003-01-17 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Gelsolin Domains 4-6 in Active, Actin-Free Conformation Identifies Sites of Regulatory Calcium Ions
J.Mol.Biol., 329, 2003
|
|
1NO6
 
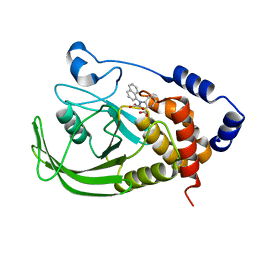 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 5 Using a Linked-Fragment Strategy | | 分子名称: | 2-[(CARBOXYCARBONYL)(1-NAPHTHYL)AMINO]BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | 登録日 | 2003-01-15 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1N8U
 
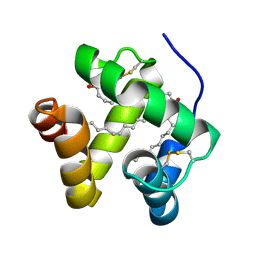 | | Chemosensory Protein in Complex with bromo-dodecanol | | 分子名称: | BROMO-DODECANOL, chemosensory protein | | 著者 | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | 登録日 | 2002-11-21 | | 公開日 | 2003-04-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NL9
 
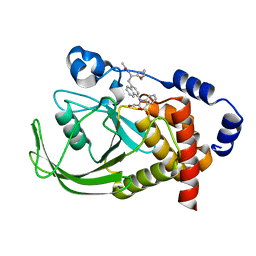 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 12 Using a Linked-Fragment Strategy | | 分子名称: | 2-{[4-(2-ACETYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-NAPHTHALEN-1-YL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | 登録日 | 2003-01-06 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of a Potent, Selective Protein Tyrosine
Phosphatase 1B Inhibitor Using a
Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1Q8D
 
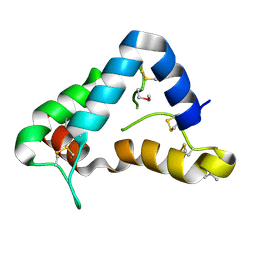 | | The crystal structure of GDNF family co-receptor alpha 1 domain 3 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GDNF family receptor alpha 1 | | 著者 | Leppanen, V.M, Bespalov, M.M, Runeberg-Roos, P, Puurand, U, Merits, A, Saarma, M, Goldman, A. | | 登録日 | 2003-08-21 | | 公開日 | 2004-08-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of GFRalpha1 domain 3 reveals new insights into GDNF binding and RET activation.
Embo J., 23, 2004
|
|
1OZF
 
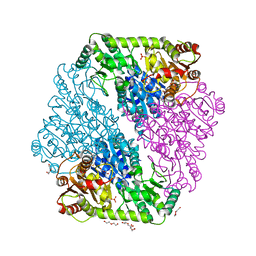 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactors | | 分子名称: | Acetolactate synthase, catabolic, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | 登録日 | 2003-04-09 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1OZH
 
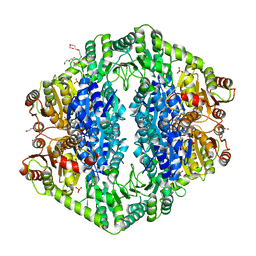 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. | | 分子名称: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | 著者 | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | 登録日 | 2003-04-09 | | 公開日 | 2003-11-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
5SRX
 
 | |
5SSI
 
 | |
5SS3
 
 | |
5SQN
 
 | |
5SS5
 
 | |
1QWD
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | 分子名称: | Outer membrane lipoprotein blc | | 著者 | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | 登録日 | 2003-09-02 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
4GGG
 
 | | Crystal structure of V66A/L68V CzrA in the Zn(II)bound state. | | 分子名称: | CHLORIDE ION, Repressor protein, ZINC ION | | 著者 | Campanello, G.C, Ma, Z, Grossoehme, N.E, Chakrovorty, D.K, Guerra, A.J, Ye, Y, Dann III, C.E, Merz Jr, K.M, Giedroc, D.P. | | 登録日 | 2012-08-06 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Allosteric inhibition of a zinc-sensing transcriptional repressor: insights into the arsenic repressor (ArsR) family.
J.Mol.Biol., 425, 2013
|
|
