3U3D
 
 | |
3TKM
 
 | | Crystal structure PPAR delta binding GW0742 | | 分子名称: | GLYCEROL, Peroxisome proliferator-activated receptor delta, {4-[({2-[3-fluoro-4-(trifluoromethyl)phenyl]-4-methyl-1,3-thiazol-5-yl}methyl)sulfanyl]-2-methylphenoxy}acetic acid | | 著者 | Trivella, D.B.B, Batista, F.H, Polikarpov, I. | | 登録日 | 2011-08-27 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Structural Insights into Human Peroxisome Proliferator Activated Receptor Delta (PPAR-Delta) Selective Ligand Binding.
Plos One, 7, 2012
|
|
1SYL
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | 著者 | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | 登録日 | 2004-04-01 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1WSG
 
 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A/D134N*) with Mn2+ | | 分子名称: | MANGANESE (II) ION, Ribonuclease HI | | 著者 | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | 登録日 | 2004-11-05 | | 公開日 | 2005-02-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
3TW2
 
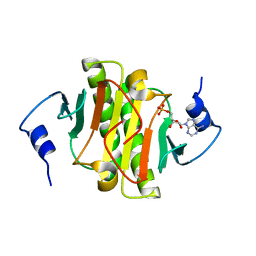 | | High resolution structure of human histidine triad nucleotide-binding protein 1 (hHINT1)/AMP complex in a monoclinic space group | | 分子名称: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | 著者 | Dolot, R.M, Wlodarczyk, A, Ozga, M, Krakowiak, A, Nawrot, B. | | 登録日 | 2011-09-21 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1WSH
 
 | |
1WTH
 
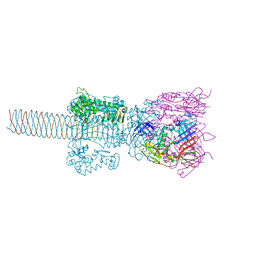 | | Crystal structure of gp5-S351L mutant and gp27 complex | | 分子名称: | Baseplate structural protein Gp27, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Kanamaru, S, Ishiwata, Y, Suzuki, T, Rossmann, M.G, Arisaka, F. | | 登録日 | 2004-11-23 | | 公開日 | 2005-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Control of bacteriophage t4 tail lysozyme activity during the infection process
J.Mol.Biol., 346, 2005
|
|
1WSI
 
 | |
3HGY
 
 | |
1WSJ
 
 | |
3R8D
 
 | |
2KHQ
 
 | | Solution NMR structure of a phage integrase SSP1947 fragment 59-159 from Staphylococcus saprophyticus, Northeast Structural Genomics Consortium Target SyR103B | | 分子名称: | Integrase | | 著者 | Eletsky, A, Mills, J.L, Hua, J, Belote, R.L, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-04-10 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of a phage integrase SSP1947 fragment 59-159 from
Staphylococcus saprophyticus
To be Published
|
|
2LWF
 
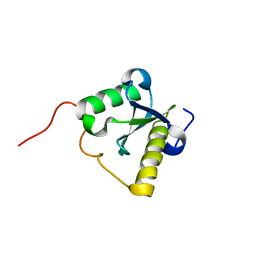 | | Structure of N-terminal domain of a plant Grx | | 分子名称: | Monothiol glutaredoxin-S16, chloroplastic | | 著者 | Feng, Y. | | 登録日 | 2012-07-28 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8VMB
 
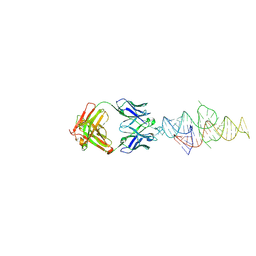 | |
8XBS
 
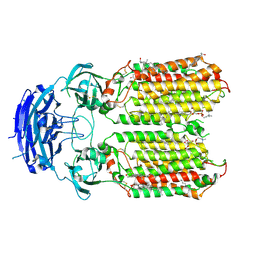 | | C. elegans apo-SID1 structure | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gong, D.S. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.21 Å) | | 主引用文献 | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8XC1
 
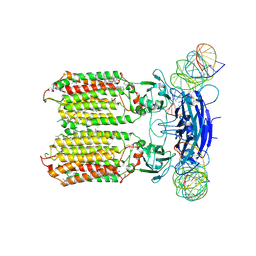 | | C. elegans SID1 in complex with dsRNA | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gong, D.S. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.21 Å) | | 主引用文献 | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
9FEH
 
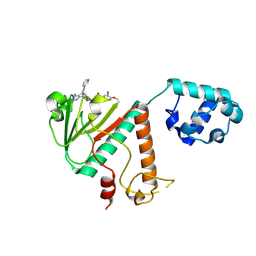 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the STM957 inhibitor | | 分子名称: | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14, ZINC ION, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-pyridin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-cyano-~{N}-ethyl-4-methoxy-benzenesulfonamide | | 著者 | Zilecka, E, Klima, M, Boura, E. | | 登録日 | 2024-05-20 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.
J Struct Biol X, 10, 2024
|
|
8W9A
 
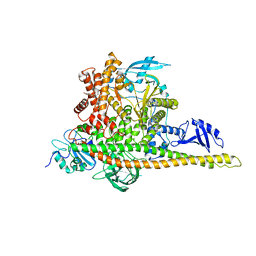 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | 分子名称: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Huang, X, Ren, X, Zhong, W. | | 登録日 | 2023-09-05 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
8VSU
 
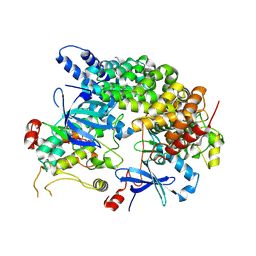 | | Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, Isoform 3 of STE20-related kinase adapter protein alpha, ... | | 著者 | Chan, L.M, Courteau, B.J, Verba, K.A. | | 登録日 | 2024-01-24 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | High-resolution single-particle imaging at 100-200 keV with the Gatan Alpine direct electron detector.
J.Struct.Biol., 216, 2024
|
|
8WMS
 
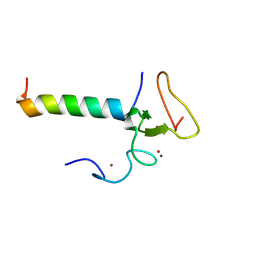 | |
8XB3
 
 | |
8VM8
 
 | |
8WIK
 
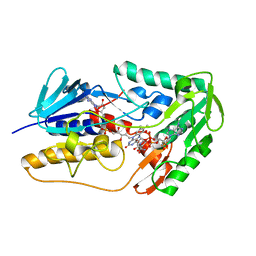 | | Crystal structure of human FSP1 | | 分子名称: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Feng, S, Huang, X, Tang, D, Qi, S. | | 登録日 | 2023-09-24 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of human ferroptosis suppressive protein 1 in complex with flavin adenine dinucleotide and nicotinamide adenine nucleotide.
MedComm (2020), 5, 2024
|
|
8XB4
 
 | |
4L59
 
 | | Crystal structure of the 3-MBT repeat domain of L3MBTL3 and UNC2533 complex | | 分子名称: | 4-(pyrrolidin-1-yl)-1-{4-[2-(pyrrolidin-1-yl)ethyl]phenyl}piperidine, Lethal(3)malignant brain tumor-like protein 3, SULFATE ION, ... | | 著者 | Zhong, N, Dong, A, Ravichandran, M, Camerino, M.A, Dickson, B.M, James, L.I, Baughman, B.M, Norris, J.L, Kireev, D.B, Janzen, W.P, Graslund, S, Frye, S.V, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-06-10 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | The structure-activity relationships of L3MBTL3 inhibitors: flexibility of the dimer interface.
Medchemcomm, 4, 2013
|
|
