8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | 分子名称: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7E
 
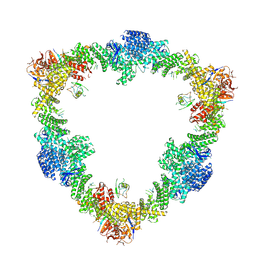 | |
8Q7H
 
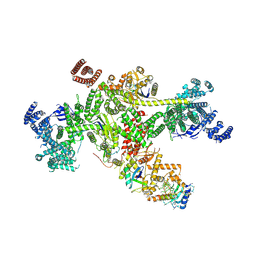 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | 分子名称: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | 著者 | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEP
 
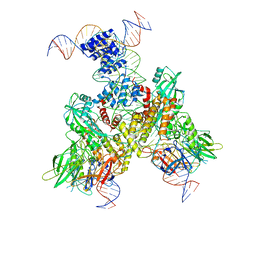 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4A0K
 
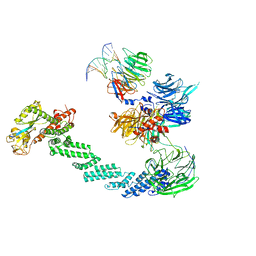 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | 分子名称: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (5.93 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
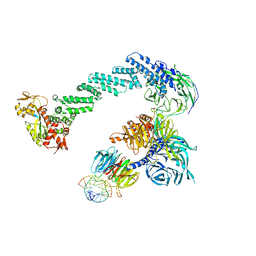 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | 分子名称: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (7.4 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
7O73
 
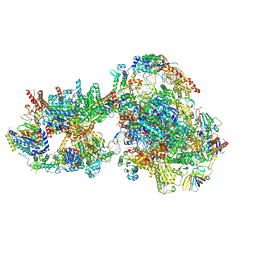 | | Yeast RNA polymerase II transcription pre-initiation complex with closed distorted promoter DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O72
 
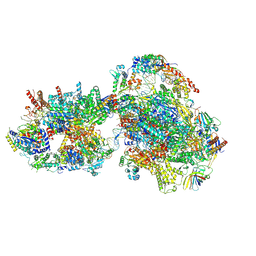 | | Yeast RNA polymerase II transcription pre-initiation complex with closed promoter DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O75
 
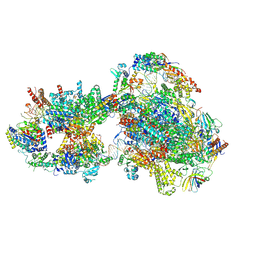 | | Yeast RNA polymerase II transcription pre-initiation complex with open promoter DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-13 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
8OO6
 
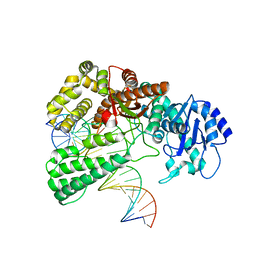 | | Pol I bound to extended and displaced DNA section - closed conformation | | 分子名称: | DNA polymerase I, Displaced primer, Extending Primer, ... | | 著者 | Botto, M, Borsellini, A, Lamers, M.H. | | 登録日 | 2023-04-04 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OOY
 
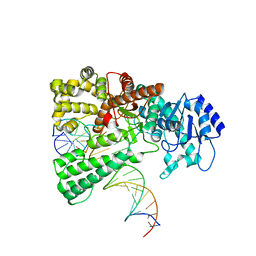 | | Pol I bound to extended and displaced DNA section - open conformation | | 分子名称: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | 著者 | Botto, M, Borsellini, A, Lamers, M.H. | | 登録日 | 2023-04-06 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2RNO
 
 | |
4E5Z
 
 | |
6R7I
 
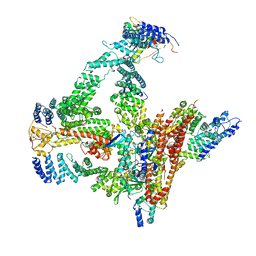 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | 登録日 | 2019-03-28 | | 公開日 | 2019-08-28 | | 最終更新日 | 2019-09-04 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
5FQD
 
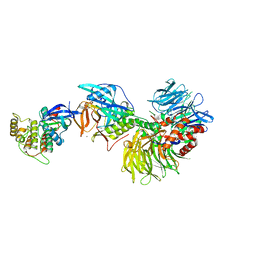 | | Structural basis of Lenalidomide induced CK1a degradation by the crl4crbn ubiquitin ligase | | 分子名称: | CASEIN KINASE I ISOFORM ALPHA, DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, ... | | 著者 | Petzold, G, Fischer, E.S, Thoma, N.H. | | 登録日 | 2015-12-09 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Basis of Lenalidomide-Induced Ck1Alpha Degradation by the Crl4(Crbn) Ubiquitin Ligase.
Nature, 532, 2016
|
|
4E54
 
 | |
2ZO1
 
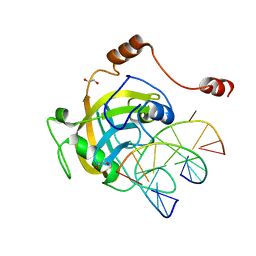 | | Mouse NP95 SRA domain DNA specific complex 2 | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), ... | | 著者 | Hashimoto, H, Horton, J.R, Cheng, X. | | 登録日 | 2008-05-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
6R7H
 
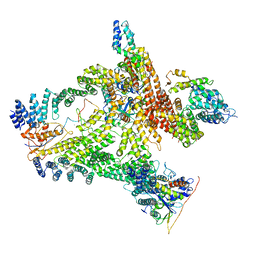 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | 登録日 | 2019-03-28 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
3EI2
 
 | |
3EI1
 
 | |
4R1M
 
 | |
4R1L
 
 | |
4RVO
 
 | |
8WQR
 
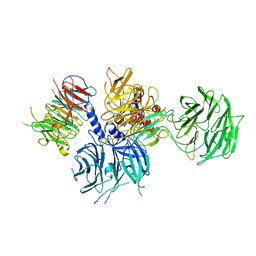 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | 分子名称: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | 著者 | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | 登録日 | 2023-10-12 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
