1PBN
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | 分子名称: | PURINE NUCLEOSIDE PHOSPHORYLASE | | 著者 | Mao, C, Ealick, S.E. | | 登録日 | 1995-07-10 | | 公開日 | 1995-11-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
6LYG
 
 | |
2BSX
 
 | | Crystal structure of the Plasmodium falciparum purine nucleoside phosphorylase complexed with inosine | | 分子名称: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | 著者 | Schnick, C, Brzozowski, A.M, Dodson, E.J, Murshudov, G.N, Brannigan, J.A, Wilkinson, A.J. | | 登録日 | 2005-05-24 | | 公開日 | 2005-08-18 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of Plasmodium Falciparum Purine Nucleoside Phosphorylase Complexed with Sulfate and its Natural Substrate Inosine
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3B9X
 
 | |
8U5H
 
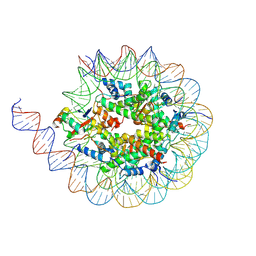 | |
4F2P
 
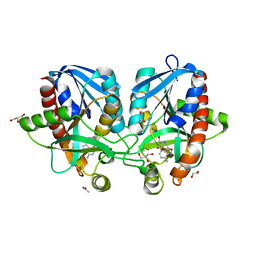 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with diEtglycol-thio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[2-(2-hydroxyethoxy)ethyl]sulfanyl}methyl)pyrrolidin-3- ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | 著者 | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2012-05-08 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with diEtglycol-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
4F3C
 
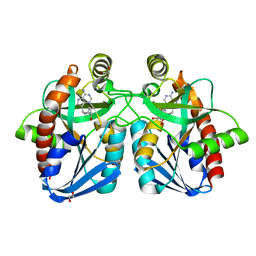 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 著者 | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2012-05-09 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
4F1W
 
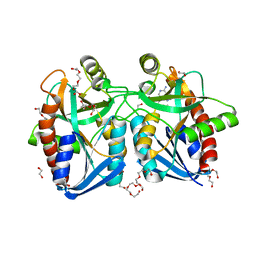 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with Adenine | | 分子名称: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | 著者 | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2012-05-07 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with Adenine
Structure, 21, 2013
|
|
4F3K
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with homocysteine-DADMe-Immucillin-A | | 分子名称: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, {(3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-hydroxypyrrolidin-3-yl}-L-methionine | | 著者 | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2012-05-09 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with homocysteine-DADMe-Immucillin-A
Structure, 21, 2013
|
|
4F2W
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with methyl-thio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | 著者 | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2012-05-08 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with methyl-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
2AI1
 
 | | Purine nucleoside phosphorylase from calf spleen | | 分子名称: | ((2R,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YL)METHYLPHOSPHONIC ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | 著者 | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | 登録日 | 2005-07-28 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AI2
 
 | | Purine nucleoside phosphorylase from calf spleen | | 分子名称: | ((2S,3AS,4R,6S)-4-(HYDROXYMETHYL)-6-(4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-TETRAHYDROFURO[3,4-D][1,3]DIOXO L-2-YL)METHYLPHOSPHONIC ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | 著者 | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | 登録日 | 2005-07-28 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4C6A
 
 | | High Resolution Structure of the Nucleoside diphosphate kinase | | 分子名称: | DI(HYDROXYETHYL)ETHER, NUCLEOSIDE DIPHOSPHATE KINASE, CYTOSOLIC | | 著者 | Priet, S, Ferron, F, Alvarez, K, Verron, M, Canard, B. | | 登録日 | 2013-09-18 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Enzymatic Synthesis of Acyclic Nucleoside Thiophosphonate Diphosphates: Effect of the Alpha-Phosphorus Configuration on HIV-1 RT Activity.
Antiviral Res., 117, 2015
|
|
2AI3
 
 | | Purine nucleoside phosphorylase from calf spleen | | 分子名称: | (2S,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YLPHOSPHONI C ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | 著者 | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | 登録日 | 2005-07-28 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A0Y
 
 | | Structure of human purine nucleoside phosphorylase H257D mutant | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Murkin, A.S, Shi, W, Schramm, V.L. | | 登録日 | 2005-06-17 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
6DYV
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-((pent-4-yn-1-ylthio)methyl)pyrrolidin-3-ol | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(pent-4-yn-1-yl)sulfanyl]methyl}pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2018-07-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
6DYU
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-((prop-2-yn-1-ylthio)methyl)pyrrolidin-3-ol | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(prop-2-yn-1-yl)sulfanyl]methyl}pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2018-07-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
6DYY
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-(((3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl)thio)methyl)pyrrolidin-3-ol | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl]sulfanyl}methyl)pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2018-07-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
6DYW
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-(((3-(1-benzyl-1H-1,2,3-triazol-4-yl)propyl)thio)methyl)pyrrolidin-3-ol | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[3-(1-benzyl-1H-1,2,3-triazol-4-yl)propyl]sulfanyl}methyl)pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | 著者 | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2018-07-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
1RR6
 
 | | Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and phosphate | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | 著者 | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | 登録日 | 2003-12-08 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function.
J.Biol.Chem., 279, 2004
|
|
2A0W
 
 | | Structure of human purine nucleoside phosphorylase H257G mutant | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Murkin, A.S, Shi, W, Schramm, V.L. | | 登録日 | 2005-06-17 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
2A0X
 
 | | Structure of human purine nucleoside phosphorylase H257F mutant | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Murkin, A.S, Shi, W, Schramm, V.L. | | 登録日 | 2005-06-17 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
5NRL
 
 | | Structure of a pre-catalytic spliceosome | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | 著者 | Plaschka, C, Lin, P.-C, Nagai, K. | | 登録日 | 2017-04-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Structure of a pre-catalytic spliceosome.
Nature, 546, 2017
|
|
4WKC
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TETRAETHYLENE GLYCOL | | 著者 | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | 登録日 | 2014-10-02 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
3B9G
 
 | | Crystal structure of loop deletion mutant of Trypanosoma vivax nucleoside hydrolase (3GTvNH) in complex with ImmH | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase, ... | | 著者 | Vandemeulebroucke, A, De Vos, S, Van Holsbeke, E, Steyaert, J, Versees, W. | | 登録日 | 2007-11-05 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A Flexible Loop as a Functional Element in the Catalytic Mechanism of Nucleoside Hydrolase from Trypanosoma vivax.
J.Biol.Chem., 283, 2008
|
|
