8E9R
 
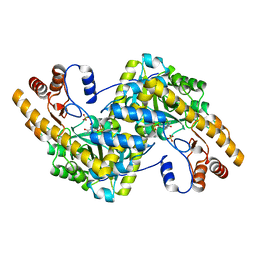 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9K
 
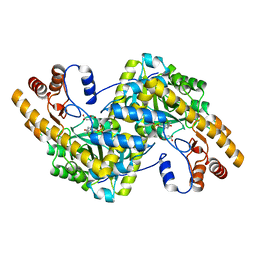 | | Crystal structure of wild-type E. coli aspartate aminotransferase bound to maleate at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9O
 
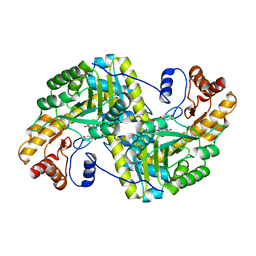 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9Q
 
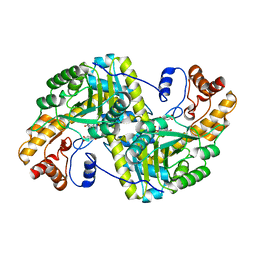 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9T
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9S
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS bound to maleic acid at 278 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9V
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
2JK6
 
 | | Structure of Trypanothione Reductase from Leishmania infantum | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TRYPANOTHIONE REDUCTASE | | 著者 | Baiocco, P, Colotti, G, Franceschini, S, Ilari, A. | | 登録日 | 2008-08-21 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Molecular Basis of Antimony Treatment in Leishmaniasis.
J.Med.Chem., 52, 2009
|
|
8E9J
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9P
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 278 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9D
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS bound to maleic acid at 100 K | | 分子名称: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9C
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS in the ligand-free form at 100 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9U
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 303 K | | 分子名称: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | 登録日 | 2022-08-26 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
4KXK
 
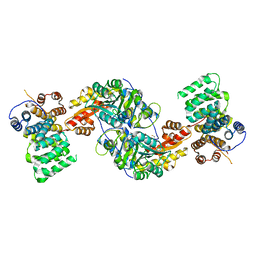 | | Alanine-glyoxylate aminotransferase variant K390A/K391A in complex with the TPR domain of human Pex5p | | 分子名称: | BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, SULFATE ION, ... | | 著者 | Fodor, K, Lou, Y, Wilmanns, M. | | 登録日 | 2013-05-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
8E77
 
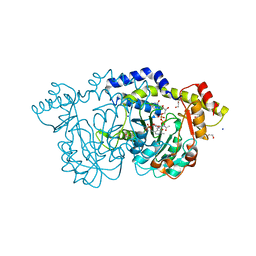 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | 分子名称: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | 著者 | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | 登録日 | 2022-08-23 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
8E75
 
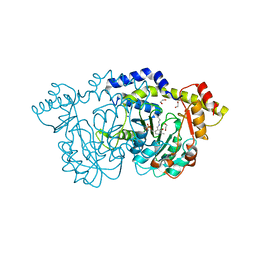 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | 分子名称: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | 著者 | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | 登録日 | 2022-08-23 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
7RFA
 
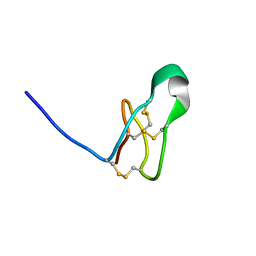 | |
4KYO
 
 | | Alanine-glyoxylate aminotransferase variant K390A in complex with the TPR domain of human Pex5p | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, ... | | 著者 | Fodor, K, Lou, Y, Wilmanns, M. | | 登録日 | 2013-05-29 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
3MY0
 
 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | 分子名称: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | 著者 | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-05-08 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
7R6O
 
 | | Pyrrolysyl-tRNA synthetase from methanogenic archaeon ISO4-G1 (G1PylRS) | | 分子名称: | 1,2-ETHANEDIOL, Pyrrolysyl-tRNA synthetase PylS, SULFATE ION | | 著者 | Frkic, R.L, Huber, T, Jackson, C.J. | | 登録日 | 2021-06-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Genetic Encoding of Cyanopyridylalanine for In-Cell Protein Macrocyclization by the Nitrile-Aminothiol Click Reaction.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5I90
 
 | |
6HIU
 
 | | Cytochrome P460 from Methylococcus capsulatus (Bath) | | 分子名称: | Cytochrome P460, GLYCEROL, HEME C, ... | | 著者 | Adams, H, Chicano, T.M, Hough, M.A. | | 登録日 | 2018-08-31 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | One fold, two functions: cytochrome P460 and cytochromec'-beta from the methanotrophMethylococcus capsulatus(Bath).
Chem Sci, 10, 2019
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | 登録日 | 2016-12-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
8FCB
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK1016790A | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, ... | | 著者 | Kwon, D.H, Lee, S.-Y, Zhang, F. | | 登録日 | 2022-12-01 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
7OH1
 
 | |
