1T6W
 
 | | RATIONAL DESIGN OF A CALCIUM-BINDING ADHESION PROTEIN NMR, 20 STRUCTURES | | 分子名称: | CALCIUM ION, hypothetical protein XP_346638 | | 著者 | Yang, W, Wilkins, A.L, Ye, Y, Liu, Z.-R, Urbauer, J.L, Kearney, A, van der Merwe, P.A, Yang, J.J. | | 登録日 | 2004-05-07 | | 公開日 | 2005-02-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of a calcium-binding protein with desired structure in a cell adhesion molecule.
J.Am.Chem.Soc., 127, 2005
|
|
3MLI
 
 | | 2ouf-ds, a disulfide-linked dimer of Helicobacter pylori protein HP0242 | | 分子名称: | CALCIUM ION, Putative uncharacterized protein | | 著者 | King, N.P, Sawaya, M.R, Jacobitz, A.W, Yeates, T.O. | | 登録日 | 2010-04-16 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and folding of a designed knotted protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | 分子名称: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | 著者 | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | 登録日 | 2014-10-27 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
8QZK
 
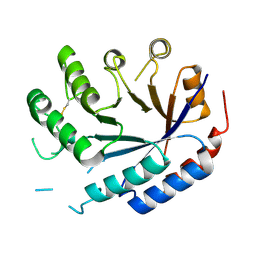 | | Catalytic core of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) concieved by deep network hallucination: dEngBF4 Hexagonal form | | 分子名称: | ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE | | 著者 | Aghajari, N, Hansen, A.L, Thiesen, F.F, Crehuet, R, Marcos, E, Willemoes, M. | | 登録日 | 2023-10-27 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Carving out a Glycoside Hydrolase Active Site for Incorporation into a New Protein Scaffold Using Deep Network Hallucination.
Acs Synth Biol, 13, 2024
|
|
2XEN
 
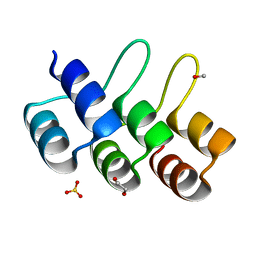 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | 分子名称: | 1,2-ETHANEDIOL, METHANOL, NI1C MUT4, ... | | 著者 | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | 登録日 | 2010-05-17 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
7RDR
 
 | |
3CWO
 
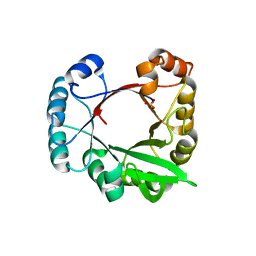 | | A beta/alpha-barrel built by the combination of fragments from different folds | | 分子名称: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | 著者 | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | 登録日 | 2008-04-22 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3MF9
 
 | |
1EKX
 
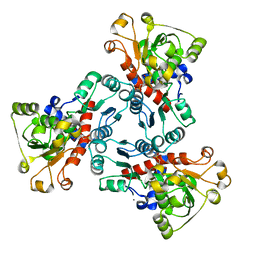 | | THE ISOLATED, UNREGULATED CATALYTIC TRIMER OF ASPARTATE TRANSCARBAMOYLASE COMPLEXED WITH BISUBSTRATE ANALOG PALA (N-(PHOSPHONACETYL)-L-ASPARTATE) | | 分子名称: | ASPARTATE TRANSCARBAMOYLASE, CALCIUM ION, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | 著者 | Endrizzi, J.A, Beernink, P.T, Alber, T, Schachman, H.K. | | 登録日 | 2000-03-09 | | 公開日 | 2000-05-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Binding of bisubstrate analog promotes large structural changes in the unregulated catalytic trimer of aspartate transcarbamoylase: implications for allosteric regulation induced cell migration.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3CSB
 
 | |
6EGP
 
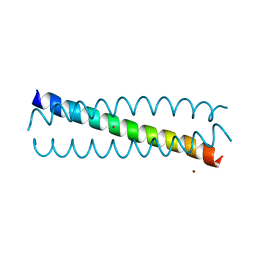 | |
8QYE
 
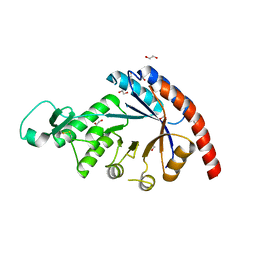 | |
7A4L
 
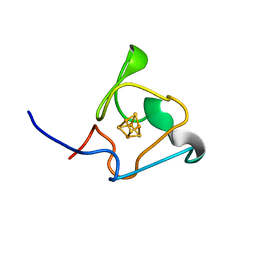 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | 分子名称: | IRON/SULFUR CLUSTER, PioC | | 著者 | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | 登録日 | 2020-08-19 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
3MFC
 
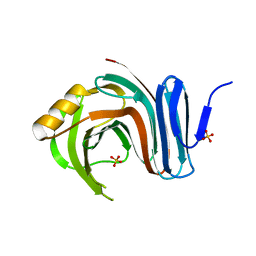 | | Computationally designed end0-1,4-beta,xylanase | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Morin, A, Harp, J.M. | | 登録日 | 2010-04-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF6
 
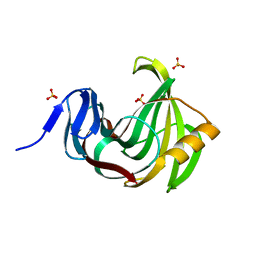 | |
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | 分子名称: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | 著者 | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | 登録日 | 2014-10-28 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
1MV4
 
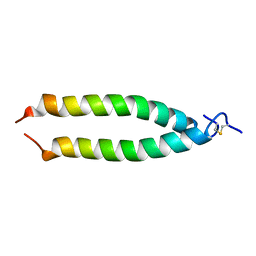 | | TM9A251-284: A Peptide Model of the C-Terminus of a Rat Striated Alpha Tropomyosin | | 分子名称: | Tropomyosin 1 alpha chain | | 著者 | Greenfield, N.J, Swapna, G.V.T, Huang, Y, Palm, T, Graboski, S, Montelione, G.T, Hitchcock-Degregori, S.E. | | 登録日 | 2002-09-24 | | 公開日 | 2003-02-18 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of the Carboxyl Terminus of Striated alpha-Tropomyosin in Solution Reveals an Unusual Parallel Arrangement of Interacting alpha-Helices
Biochemistry, 42, 2003
|
|
8V2D
 
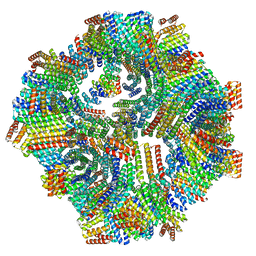 | |
8VC8
 
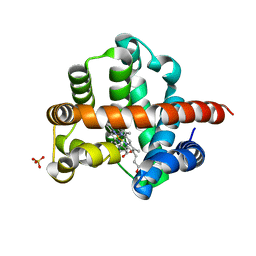 | |
4NEY
 
 | | Crystal Structure of an engineered protein with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR277 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Engineered protein OR277, tetrabutylphosphonium | | 著者 | Guan, R, Lin, Y.-R, Koga, N, Koga, R, Castellanos, J, Seetharaman, J, Maglaqui, M, Sahdev, S, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-10-30 | | 公開日 | 2014-01-15 | | 実験手法 | X-RAY DIFFRACTION (2.323 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target OR277
To be published
|
|
4NEZ
 
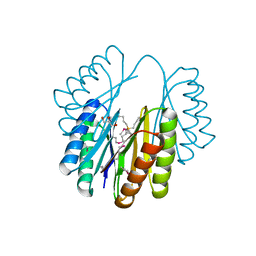 | | Crystal Structure of an engineered protein with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR276 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Engineered protein OR276, tetrabutylphosphonium | | 著者 | Guan, R, Lin, Y.-R, Koga, N, Koga, R, Castellanos, J, Seetharaman, J, Maglaqui, M, Sahdev, S, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-10-30 | | 公開日 | 2014-01-15 | | 実験手法 | X-RAY DIFFRACTION (2.395 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target OR276
To be published
|
|
6REG
 
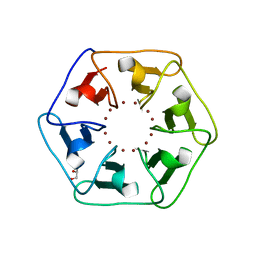 | | Crystal structure of Pizza6-S with Zn2+ | | 分子名称: | GLYCEROL, Pizza6-S, ZINC ION | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REK
 
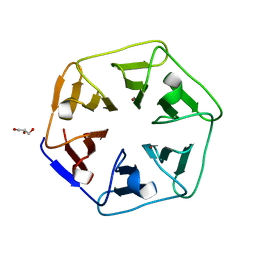 | | Crystal structure of Pizza6-SH with Cu2+ | | 分子名称: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REN
 
 | | Crystal structure of 3fPizza6-SH with Zn2+ | | 分子名称: | 3fPizza6-SH, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REH
 
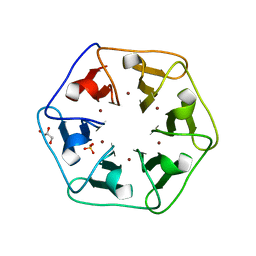 | | Crystal structure of Pizza6-S with Cu2+ | | 分子名称: | COPPER (II) ION, GLYCEROL, Pizza6-S, ... | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
