7OML
 
 | |
9D5Z
 
 | | Crystal structure of the human WDR5 in complex with LH168 compound | | 分子名称: | (3P)-N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[1-ethyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]-5-[(1H-imidazol-1-yl)methyl]benzamide, 1,2-ETHANEDIOL, WD repeat-containing protein 5 | | 著者 | Kimani, S, Dong, A, Hoffmann, L, Nemec, V, Ackloo, S, Muller-Knapp, S, Knapp, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2024-08-14 | | 公開日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the human WDR5 in complex with LH168 compound
To be published
|
|
6OG0
 
 | | Structure of Aedes aegypti OBP22 | | 分子名称: | AAEL005772-PA, CADMIUM ION, CHLORIDE ION | | 著者 | Jones, D.N, Wang, J. | | 登録日 | 2019-04-01 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Aedes aegypti Odorant Binding Protein 22 selectively binds fatty acids through a conformational change in its C-terminal tail.
Sci Rep, 10, 2020
|
|
7TGG
 
 | | Cryo-EM structure of PilA-N and PilA-C from Geobacter sulfurreducens | | 分子名称: | Geopilin domain 1 protein, Geopilin domain 2 protein | | 著者 | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Bond, D.R, Hochbaum, A.I, Egelman, E.H. | | 登録日 | 2022-01-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
6Y2X
 
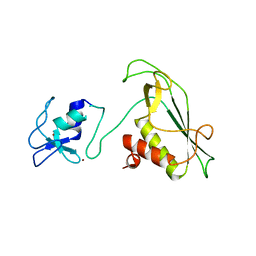 | |
6OII
 
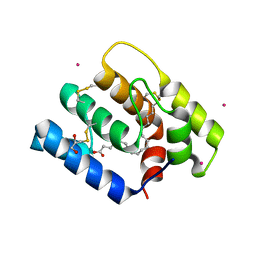 | |
7ZJY
 
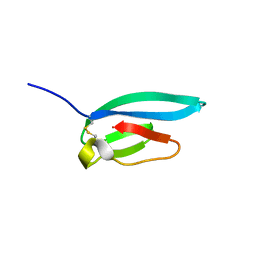 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | 分子名称: | MAX effector protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZK0
 
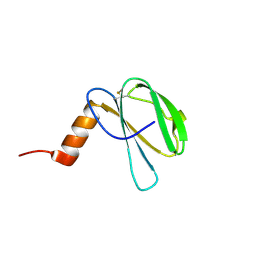 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | 分子名称: | MAX effector protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZKD
 
 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | 分子名称: | MAX effector protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7SNN
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide | | 分子名称: | CHLORIDE ION, Capsid protein p24, IODIDE ION, ... | | 著者 | Bester, S.M, Kvaratskhelia, M. | | 登録日 | 2021-10-28 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor N-(1-(3-(4-chloro-1-methyl-3-(methylsulfonamido)-1H-indazol-7-yl)-4-oxo-3,4-dihydropyrido[2,3-d]pyrimidin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(difluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl)acetamide
To Be Published
|
|
8T28
 
 | | The crystal structure of SrtC2 sortase from Actinomyces oris | | 分子名称: | CHLORIDE ION, Class C sortase, PHOSPHATE ION | | 著者 | Osipiuk, J, Chang, C, Ton-That, H.L, Ton-That, H, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-06-05 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis for dual functions in pilus assembly modulated by the lid of a pilus-specific sortase.
J.Biol.Chem., 300, 2024
|
|
8BCX
 
 | |
6OPB
 
 | |
4WH8
 
 | | Crystal Structure of HCV NS3/4A protease in complex with an Asunaprevir P1-P3 macrocyclic analog. | | 分子名称: | Genome polyprotein, SULFATE ION, ZINC ION, ... | | 著者 | Soumana, D.I, Ali, A, Schiffer, C.A. | | 登録日 | 2014-09-20 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
4WF8
 
 | | Crystal structure of NS3/4A protease in complex with Asunaprevir | | 分子名称: | CHLORIDE ION, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, NS3 protein, ... | | 著者 | Schiffer, C.A, Soumana, D.I, Ali, A. | | 登録日 | 2014-09-13 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
8P9I
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB462 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9H
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB437 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-2-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)quinoline-6-carboxamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9K
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB503 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9J
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB500 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9L
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
6WXZ
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-29 A.K.A 7-(1,2-DIPHENYLETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1,2-diphenylethyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | 著者 | Khan, J.A. | | 登録日 | 2020-05-12 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6PIV
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | 分子名称: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2019-06-27 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PJ0
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | 分子名称: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2019-06-27 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
8B1O
 
 | |
8B1P
 
 | |
