8G32
 
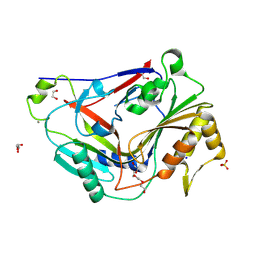 | | Pro-form of a CDCL short from E. anophelis | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Johnstone, B.A, Christie, M.P, Morton, C.J, Parker, M.W. | | 登録日 | 2023-02-06 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Distant relatives of a eukaryotic cell-specific toxin family evolved a complement-like mechanism to kill bacteria.
Nat Commun, 15, 2024
|
|
7AVW
 
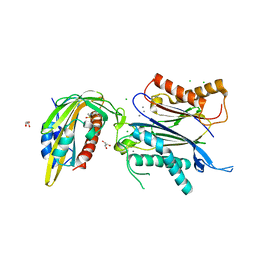 | |
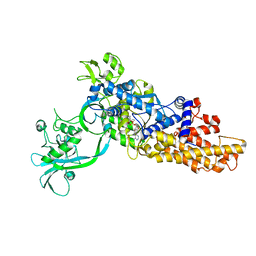
6LDK
 
 | |
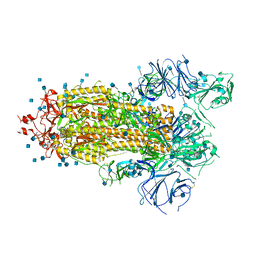
7KDH
 
 | |
7KEB
 
 | |
7ZW6
 
 | | Oligomeric structure of SynDLP | | 分子名称: | Slr0869 protein | | 著者 | Gewehr, L, Junglas, B, Jilly, R, Franz, J, Wenyu, E.Z, Weidner, T, Bonn, M, Sachse, C, Schneider, D. | | 登録日 | 2022-05-18 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | SynDLP is a dynamin-like protein of Synechocystis sp. PCC 6803 with eukaryotic features.
Nat Commun, 14, 2023
|
|
7KDI
 
 | |
7KEA
 
 | |
7TDR
 
 | |
7TDS
 
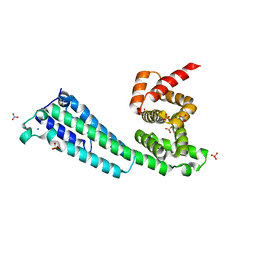 | |
7KE9
 
 | |
7K16
 
 | | Tamana Bat Virus xrRNA1 | | 分子名称: | MAGNESIUM ION, RNA (51-MER), SODIUM ION | | 著者 | Jones, R.A, Kieft, J.S. | | 登録日 | 2020-09-07 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Different tertiary interactions create the same important 3D features in a distinct flavivirus xrRNA.
Rna, 27, 2021
|
|
7KDG
 
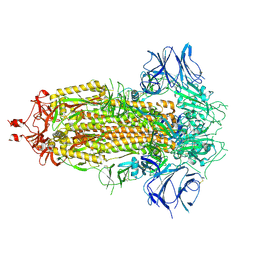 | |
7KDL
 
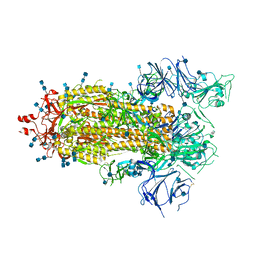 | |
7KDJ
 
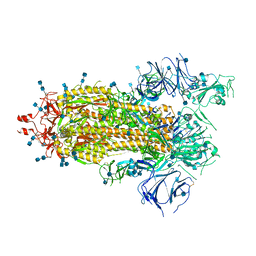 | |
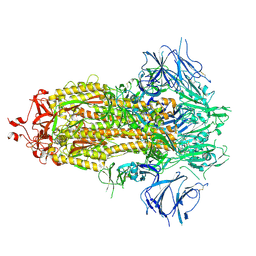
7KE4
 
 | |
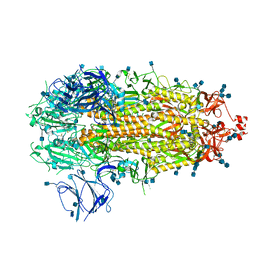
7KDK
 
 | |
7KEC
 
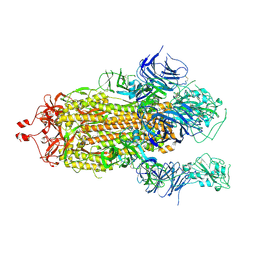 | |
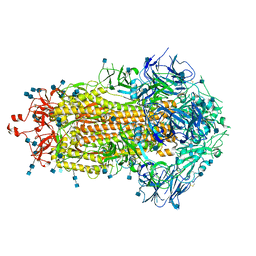
7KE7
 
 | |
7KE6
 
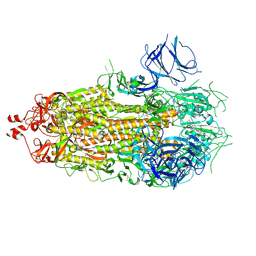 | |
3WCU
 
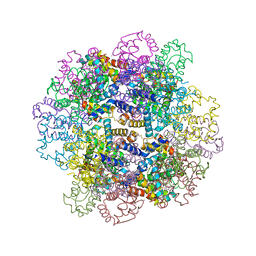 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Deoxygenated form | | 分子名称: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | 著者 | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | 登録日 | 2013-06-01 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCV
 
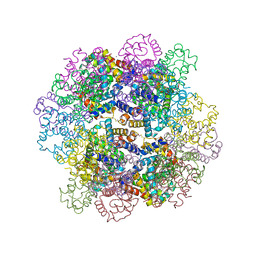 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: CA bound form | | 分子名称: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | 著者 | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | 登録日 | 2013-06-01 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCW
 
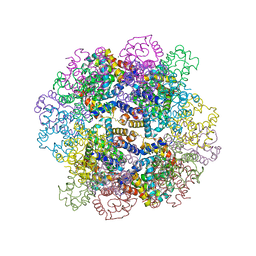 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: MG bound form | | 分子名称: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | 著者 | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | 登録日 | 2013-06-01 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCT
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Oxygenated form | | 分子名称: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | 著者 | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | 登録日 | 2013-06-01 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8BZS
 
 | | The crystal structure of glycogen phosphorylase in complex with baicalein | | 分子名称: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Glycogen phosphorylase, muscle form, ... | | 著者 | Koulas, S.M, Mathomes, R, Hayes, J.M, Leonidas, D.D. | | 登録日 | 2022-12-15 | | 公開日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Multidisciplinary docking, kinetics and X-ray crystallography studies of baicalein acting as a glycogen phosphorylase inhibitor and determination of its' potential against glioblastoma in cellular models.
Chem.Biol.Interact., 382, 2023
|
|
