8FC9
 
 | | Cryo-EM structure of the human TRPV4 - RhoA, apo condition | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA, ... | | 著者 | Kwon, D.H, Lee, S.-Y, Zhang, F. | | 登録日 | 2022-12-01 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FCM
 
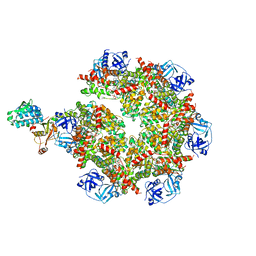 | | Cryo-EM structure of p97:UBXD1 open state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FC8
 
 | | Cryo-EM structure of the human TRPV4 in complex with GSK1016790A | | 分子名称: | CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | 著者 | Kwon, D.H, Lee, S.-Y, Zhang, F. | | 登録日 | 2022-12-01 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FCB
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK1016790A | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, ... | | 著者 | Kwon, D.H, Lee, S.-Y, Zhang, F. | | 登録日 | 2022-12-01 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FCA
 
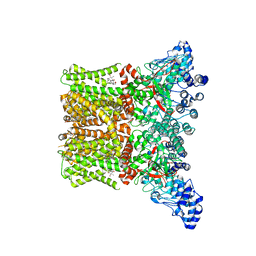 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with 4alpha-Phorbol 12,13-didecanoate | | 分子名称: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily V member 4 | | 著者 | Kwon, D.H, Lee, S.-Y, Zhang, F. | | 登録日 | 2022-12-01 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FCO
 
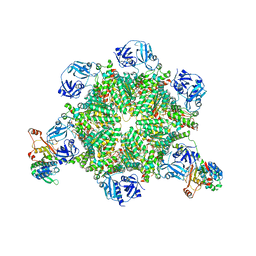 | | Cryo-EM structure of p97:UBXD1 meta state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCN
 
 | | Cryo-EM structure of p97:UBXD1 VIM-only state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCP
 
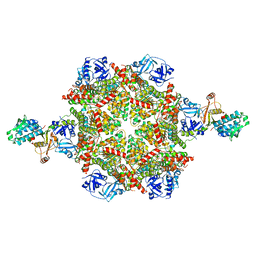 | | Cryo-EM structure of p97:UBXD1 para state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCL
 
 | | Cryo-EM structure of p97:UBXD1 closed state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HLP
 
 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Wei, Y, Yu, Z, Zhao, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8BUU
 
 | | ARE-ABCF VmlR2 bound to a 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Crowe-McAuliffe, C, Wilson, D.N. | | 登録日 | 2022-11-30 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR.
Nucleic Acids Res., 51, 2023
|
|
8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | 分子名称: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU7
 
 | | Structure of DDB1 bound to 21195-engaged CDK12-cyclin K | | 分子名称: | 1,2-ETHANEDIOL, 1-[2,6-bis(chloranyl)phenyl]-6-[[4-(2-hydroxyethyloxy)phenyl]methyl]-3-propan-2-yl-5H-pyrazolo[3,4-d]pyrimidin-4-one, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.245 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUN
 
 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUA
 
 | | Structure of DDB1 bound to 919278-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-~{N}-(1~{H}-benzimidazol-2-yl)-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.193 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU2
 
 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUR
 
 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUI
 
 | | Structure of DDB1 bound to DRF-053-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)amino]purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUF
 
 | | Structure of DDB1 bound to Z12-engaged CDK12-cyclin K | | 分子名称: | 2-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylmethyl)-6,7-dimethoxy-3~{H}-quinazolin-4-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU1
 
 | | Structure of DDB1 bound to DS17-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUO
 
 | | Structure of DDB1 bound to DS24-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(3-fluoranyl-4-pyridin-2-yl-phenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.58 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU4
 
 | | Structure of DDB1 bound to DS22-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU6
 
 | | Structure of DDB1 bound to DS55-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUJ
 
 | | Structure of DDB1 bound to DS06-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-(octylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.62 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUK
 
 | | Structure of DDB1 bound to DS08-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-(naphthalen-2-ylmethylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
