6G6U
 
 | |
6PHT
 
 | |
6PID
 
 | |
6PI8
 
 | |
6PHZ
 
 | | Crystal structure of Marinobacter subterrani acetylpolyamine amidohydrolase (msAPAH) complexed with 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one | | 分子名称: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, Acetylpolyamine Amidohydrolase, MAGNESIUM ION, ... | | 著者 | Osko, J.D, Christianson, D.W. | | 登録日 | 2019-06-25 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Function of the Acetylpolyamine Amidohydrolase from the Deep Earth HalophileMarinobacter subterrani.
Biochemistry, 58, 2019
|
|
6PIA
 
 | |
8JDM
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (rotated state) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDL
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (non-rotated state) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDJ
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDK
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
5L4O
 
 | |
8YJF
 
 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and an H2A-H2B dimer | | 分子名称: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2A type 1-D, ... | | 著者 | Gan, S.L, Yang, W.S, Xu, R.M. | | 登録日 | 2024-03-01 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
4GEN
 
 | | Crystal structure of Zucchini (monomer) | | 分子名称: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | 著者 | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEM
 
 | | Crystal structure of Zucchini (K171A) | | 分子名称: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | 著者 | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
5MIY
 
 | | Crystal structure of the E3 ubiquitin ligase RavN from Legionella pneumophila | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin ligase RavN, SODIUM ION, ... | | 著者 | Lucas, M, Abascal-Palacios, G, Rojas, A.L, Hierro, A. | | 登録日 | 2016-11-29 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.717 Å) | | 主引用文献 | RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases.
PLoS Pathog., 14, 2018
|
|
5A1Y
 
 | | The structure of the COPI coat linkage IV | | 分子名称: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | 著者 | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | 登録日 | 2015-05-06 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (21 Å) | | 主引用文献 | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
8HY0
 
 | |
8HXZ
 
 | |
8HXY
 
 | |
5A1V
 
 | | The structure of the COPI coat linkage I | | 分子名称: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | 著者 | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | 登録日 | 2015-05-06 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (21 Å) | | 主引用文献 | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
1NKF
 
 | |
8I7R
 
 | | In situ structure of axonemal doublet microtubules in mouse sperm with 48-nm repeat | | 分子名称: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | 著者 | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | 登録日 | 2023-02-02 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
6D1V
 
 | |
1NQC
 
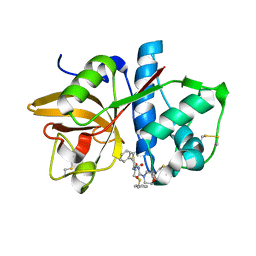 | | Crystal structures of Cathepsin S inhibitor complexes | | 分子名称: | Cathepsin S, N-[(1R)-2-(BENZYLSULFANYL)-1-FORMYLETHYL]-N-(MORPHOLIN-4-YLCARBONYL)-L-PHENYLALANINAMIDE | | 著者 | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Menard, R, Cygler, M, Rath, V.L. | | 登録日 | 2003-01-21 | | 公開日 | 2003-04-15 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
1OED
 
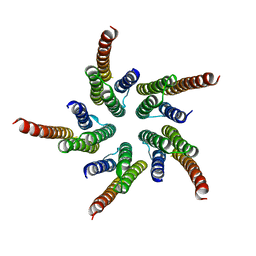 | | STRUCTURE OF ACETYLCHOLINE RECEPTOR PORE FROM ELECTRON IMAGES | | 分子名称: | Acetylcholine receptor beta subunit, Acetylcholine receptor delta subunit, Acetylcholine receptor gamma subunit, ... | | 著者 | Miyazawa, A, Fujiyoshi, Y, Unwin, N. | | 登録日 | 2003-03-24 | | 公開日 | 2003-06-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure and Gating Mechanism of the Acetylcholine Receptor Pore.
Nature, 423, 2003
|
|
