5NDH
 
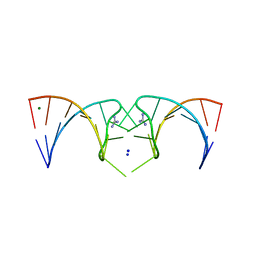 | | The structure of the G. violaceus guanidine II riboswitch P2 stem-loop | | 分子名称: | GUANIDINE, MAGNESIUM ION, RNA (5'-R(*GP*(CBV)P*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*GP*C)-3'), ... | | 著者 | Huang, L, Wang, J, Lilley, D.M.J. | | 登録日 | 2017-03-08 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NEP
 
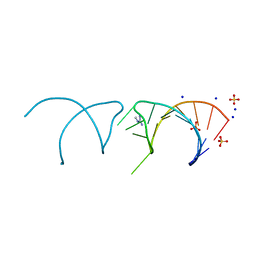 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with methylguanidine | | 分子名称: | 1-METHYLGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | 著者 | Huang, L, Wang, J, Lilley, D.M.J. | | 登録日 | 2017-03-11 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NEO
 
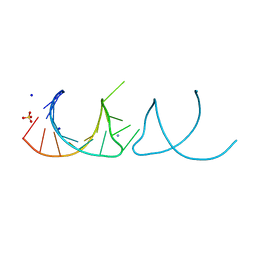 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop | | 分子名称: | AMMONIUM ION, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | 著者 | Huang, L, Wang, J, Lilley, D.M.J. | | 登録日 | 2017-03-11 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NEQ
 
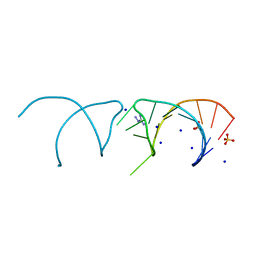 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with aminoguanidine | | 分子名称: | AMINOGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | 著者 | Huang, L, Wang, J, Lilley, D.M.J. | | 登録日 | 2017-03-11 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NEX
 
 | |
5NOM
 
 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with guanidine | | 分子名称: | GUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | 著者 | Huang, L, Wang, J, Lilley, D.M.J. | | 登録日 | 2017-04-12 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NDI
 
 | |
8HB1
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | 著者 | Peng, X, Lilley, D.M.J, Huang, L. | | 登録日 | 2022-10-27 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
3MUT
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Smith, K.D. | | 登録日 | 2010-05-03 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUR
 
 | | Crystal Structure of the C92U mutant c-di-GMP riboswith bound to c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Smith, K.D. | | 登録日 | 2010-05-03 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUM
 
 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Smith, K.D. | | 登録日 | 2010-05-03 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUV
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Smith, K.D. | | 登録日 | 2010-05-03 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
6TF0
 
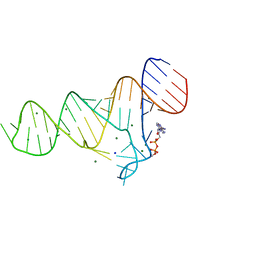 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2019-11-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TB7
 
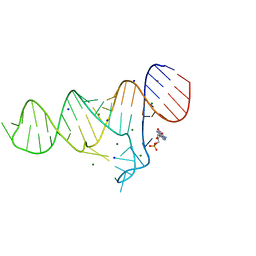 | |
6TF1
 
 | |
3K0J
 
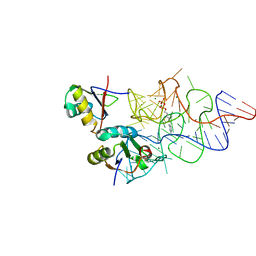 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | 分子名称: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | 著者 | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | 登録日 | 2009-09-24 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
6C27
 
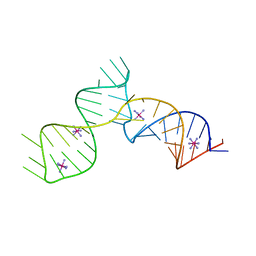 | | SAM-III riboswitch ON-state | | 分子名称: | COBALT HEXAMMINE(III), SAM-III riboswitch | | 著者 | Grigg, J.C, Price, I.R, Ke, A. | | 登録日 | 2018-01-07 | | 公開日 | 2019-01-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.601 Å) | | 主引用文献 | Evidence for two-tiered conformation selection in the SAM-III riboswitch
To Be Published
|
|
6WJS
 
 | |
6WJR
 
 | |
3DIM
 
 | |
3DJ2
 
 | |
3GX5
 
 | |
3GX6
 
 | |
3DIY
 
 | |
3DIO
 
 | |
