2IV2
 
 | | Reinterpretation of reduced form of formate dehydrogenase H from E. coli | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase H, GUANYLATE-O'-PHOSPHORIC ACID MONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,5,6,7,8A,9,10,10A-OCTAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL) ESTER, ... | | 著者 | Raaijmakers, H.C.A, Romao, M.J. | | 登録日 | 2006-06-08 | | 公開日 | 2006-06-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Formate-Reduced E. Coli Formate Dehydrogenase H: The Reinterpretation of the Crystal Structure Suggests a New Reaction Mechanism.
J.Biol.Inorg.Chem., 11, 2006
|
|
1ED9
 
 | | STRUCTURE OF E. COLI ALKALINE PHOSPHATASE WITHOUT THE INORGANIC PHOSPHATE AT 1.75A RESOLUTION | | 分子名称: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Stec, B, Holtz, K.M, Kantrowitz, E.R. | | 登録日 | 2000-01-27 | | 公開日 | 2000-09-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A revised mechanism for the alkaline phosphatase reaction involving three metal ions.
J.Mol.Biol., 299, 2000
|
|
3HVJ
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-propyladenine-containing bisubstrate inhibitor | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | 著者 | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | 登録日 | 2009-06-16 | | 公開日 | 2009-10-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1B9Z
 
 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | 分子名称: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | 著者 | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | 登録日 | 1999-03-06 | | 公開日 | 1999-03-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
8TFU
 
 | |
8TGC
 
 | |
5I3H
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, POTASSIUM ION, Triosephosphate isomerase, ... | | 著者 | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | 登録日 | 2016-02-10 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | 分子名称: | O-GLCNACASE NAGJ | | 著者 | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | 登録日 | 2007-02-19 | | 公開日 | 2007-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
5I3F
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | 分子名称: | Triosephosphate isomerase, glycosomal | | 著者 | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | 登録日 | 2016-02-10 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
3HVK
 
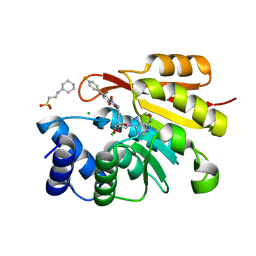 | | Rat catechol O-methyltransferase in complex with a catechol-type, purine-containing bisubstrate inhibitor - humanized form | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | 著者 | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | 登録日 | 2009-06-16 | | 公開日 | 2009-10-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
7MCH
 
 | |
1BEU
 
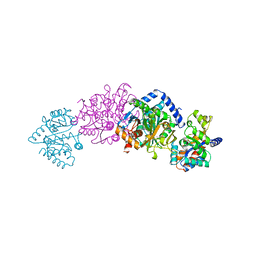 | | TRP SYNTHASE (D60N-IPP-SER) WITH K+ | | 分子名称: | INDOLE-3-PROPANOL PHOSPHATE, POTASSIUM ION, TRYPTOPHAN SYNTHASE, ... | | 著者 | Rhee, S, Mozzarelli, A, Miles, E.W, Davies, D.R. | | 登録日 | 1998-05-18 | | 公開日 | 1998-08-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cryocrystallography and microspectrophotometry of a mutant (alpha D60N) tryptophan synthase alpha 2 beta 2 complex reveals allosteric roles of alpha Asp60.
Biochemistry, 37, 1998
|
|
3I4N
 
 | | 8-oxoguanine containing RNA polymerase II elongation complex E | | 分子名称: | DNA (5'-D(*AP*G*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*(8OG)P*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Damsma, G.E, Cramer, P. | | 登録日 | 2009-07-02 | | 公開日 | 2009-09-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Molecular basis of transcriptional mutagenesis at 8-oxoguanine
J.Biol.Chem., 284, 2009
|
|
1EW8
 
 | | ALKALINE PHOSPHATASE (E.C. 3.1.3.1) COMPLEX WITH PHOSPHONOACETIC ACID | | 分子名称: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Holtz, K.M, Stec, B, Myers, J.K, Antonelli, S.M, Widlanski, T.S, Kantrowitz, E.R. | | 登録日 | 2000-04-24 | | 公開日 | 2002-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Alternate modes of binding in two crystal structures of alkaline phosphatase-inhibitor complexes.
Protein Sci., 9, 2000
|
|
3IJI
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; nonproductive substrate binding. | | 分子名称: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-08-04 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5IR5
 
 | | Crystal structure of wild-type bacterial lipoxygenase from Pseudomonas aeruginosa PA-LOX with space group P21212 at 1.9 A resolution | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, Arachidonate 15-lipoxygenase, FE (II) ION, ... | | 著者 | Kalms, J, Banthiya, S, Galemou Yoga, E, Kuhn, H, Scheerer, P. | | 登録日 | 2016-03-12 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa.
Biochim.Biophys.Acta, 1861, 2016
|
|
1PD5
 
 | |
2SCU
 
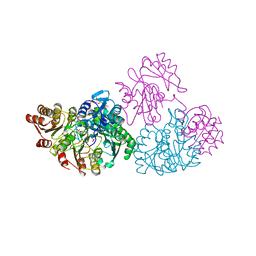 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | 分子名称: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | 著者 | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | 登録日 | 1998-09-24 | | 公開日 | 1999-08-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
5I3J
 
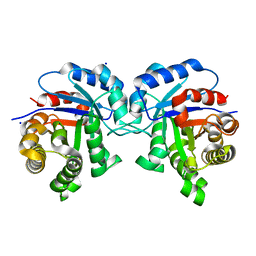 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | 分子名称: | SODIUM ION, Triosephosphate isomerase, glycosomal | | 著者 | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | 登録日 | 2016-02-10 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
7SHX
 
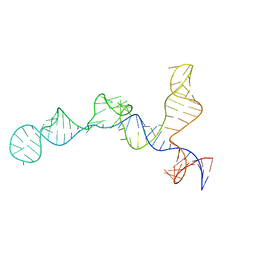 | |
3IJQ
 
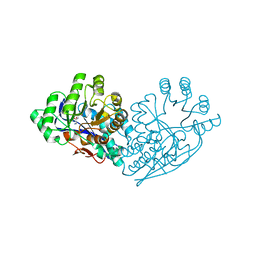 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | 分子名称: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-08-04 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OWD
 
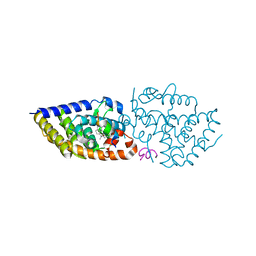 | | Vitamin D receptor complex | | 分子名称: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-hept-5-en-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N, Li, W. | | 登録日 | 2017-08-31 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
1BOT
 
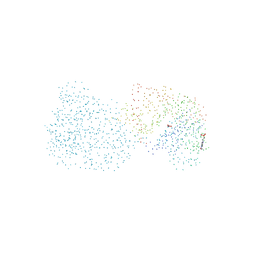 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | 著者 | Ormo, M, Bystrom, C.E, Remington, S.J. | | 登録日 | 1998-08-05 | | 公開日 | 1999-01-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
5I3K
 
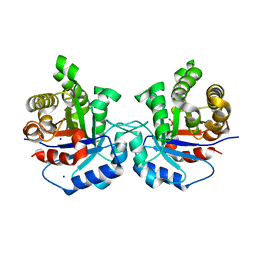 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, SODIUM ION, Triosephosphate isomerase, ... | | 著者 | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | 登録日 | 2016-02-10 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.209 Å) | | 主引用文献 | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5IFG
 
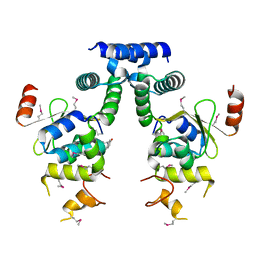 | | Crystal structure of HigA-HigB complex from E. Coli | | 分子名称: | Antitoxin HigA, mRNA interferase HigB | | 著者 | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | 登録日 | 2016-02-26 | | 公開日 | 2017-03-01 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
