1GEI
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | 分子名称: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | 登録日 | 2000-11-13 | | 公開日 | 2000-11-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
1L30
 
 | |
4IK3
 
 | | High resolution structure of GCaMP3 at pH 8.5 | | 分子名称: | CALCIUM ION, RCaMP, Green fluorescent protein | | 著者 | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | 登録日 | 2012-12-25 | | 公開日 | 2014-01-29 | | 最終更新日 | 2017-06-21 | | 実験手法 | X-RAY DIFFRACTION (2.007 Å) | | 主引用文献 | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
1GF9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1LGW
 
 | | T4 Lysozyme Mutant L99A/M102Q Bound by 2-fluoroaniline | | 分子名称: | 2-FLUOROANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1GFJ
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1LHM
 
 | |
3BG1
 
 | |
1LHZ
 
 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 293K | | 分子名称: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | 著者 | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | 登録日 | 2002-04-17 | | 公開日 | 2003-07-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
1L36
 
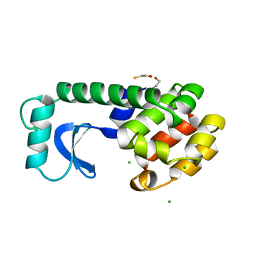 | |
1L3H
 
 | | NMR structure of P41icf, a potent inhibitor of human cathepsin L | | 分子名称: | MHC CLASS II-ASSOCIATED P41 INVARIANT CHAIN FRAGMENT (P41icf) | | 著者 | Chiva, C, Barthe, P, Codina, A, Giralt, E. | | 登録日 | 2002-02-27 | | 公開日 | 2003-03-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
J.Am.Chem.Soc., 125, 2003
|
|
1LIR
 
 | | LQ2 FROM LEIURUS QUINQUESTRIATUS, NMR, 22 STRUCTURES | | 分子名称: | LQ2 | | 著者 | Renisio, J.G, Lu, Z, Blanc, E, Jin, W, Lewis, J.H, Bornet, O, Darbon, H. | | 登録日 | 1998-04-02 | | 公開日 | 1998-06-17 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of potassium channel-inhibiting scorpion toxin Lq2.
Proteins, 34, 1999
|
|
1LJN
 
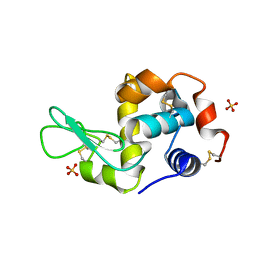 | |
4TM4
 
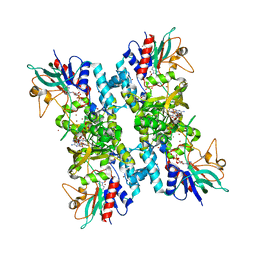 | |
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | 分子名称: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | 著者 | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | 登録日 | 2000-12-07 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
1LL4
 
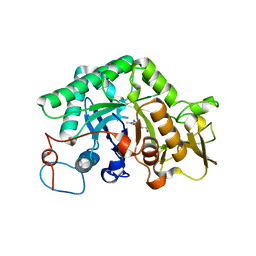 | | STRUCTURE OF C. IMMITIS CHITINASE 1 COMPLEXED WITH ALLOSAMIDIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE 1 | | 著者 | Bortone, K, Monzingo, A.F, Ernst, S, Robertus, J.D. | | 登録日 | 2002-04-26 | | 公開日 | 2002-09-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | THE STRUCTURE OF AN ALLOSAMIDIN COMPLEX WITH THE Coccidioides IMMITIS CHITINASE DEFINES A ROLE FOR A SECOND ACID RESIDUE IN SUBSTRATE-ASSISTED MECHANISM
J.Mol.Biol., 320, 2002
|
|
1L52
 
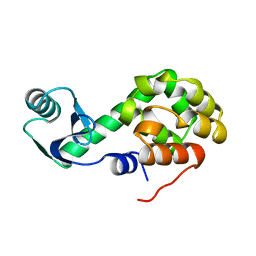 | |
1LLZ
 
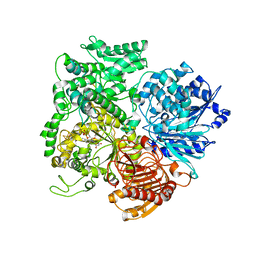 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: reduced enzyme | | 分子名称: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, Ferredoxin-dependent glutamate synthase | | 著者 | van den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | 登録日 | 2002-04-30 | | 公開日 | 2002-07-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
6GNZ
 
 | |
2AK1
 
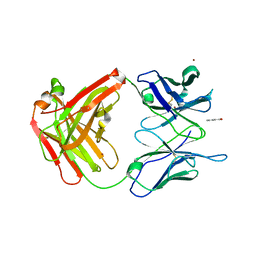 | |
4TSZ
 
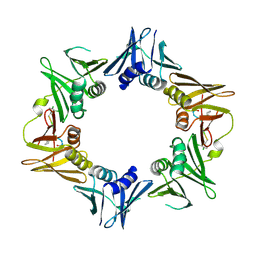 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | 分子名称: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-19 | | 公開日 | 2014-09-10 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
1L77
 
 | |
4AFT
 
 | | Aplysia californica AChBP in complex with Varenicline | | 分子名称: | SOLUBLE ACETYLCHOLINE RECEPTOR, VARENICLINE | | 著者 | Rucktooa, P, Haseler, C.A, vanElke, R, Smit, A.B, Gallagher, T, Sixma, T.K. | | 登録日 | 2012-01-23 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Characterization of Binding Mode of Smoking Cessation Drugs to Nicotinic Acetylcholine Receptors Through Study of Ligand Complexes with Acetylcholine-Binding Protein.
J.Biol.Chem., 287, 2012
|
|
6GO0
 
 | |
2AJN
 
 | | NMR structure of the in-plane membrane anchor domain [1-28] of the monotopic NonStructural Protein 5A (NS5A) from the Bovine Viral Diarrhea Virus (BVDV) | | 分子名称: | Nonstructural protein 5A | | 著者 | Sapay, N, Montserret, R, Chipot, C, Brass, V, Moradpour, D, Deleage, G, Penin, F. | | 登録日 | 2005-08-02 | | 公開日 | 2005-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus.
Biochemistry, 45, 2006
|
|
