1YSF
 
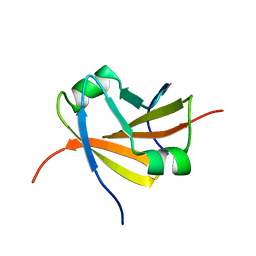 | |
1BO8
 
 | | THYMIDYLATE SYNTHASE R178T MUTANT | | 分子名称: | POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE), URIDINE-5'-MONOPHOSPHATE | | 著者 | Morse, R, Finer-Moore, J, Stroud, R.M. | | 登録日 | 1998-08-10 | | 公開日 | 1998-08-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1H7O
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 5-AMINOLAEVULINIC ACID AT 1.7 A RESOLUTION | | 分子名称: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, DELTA-AMINO VALERIC ACID, ZINC ION | | 著者 | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | 登録日 | 2001-07-09 | | 公開日 | 2001-07-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1BWH
 
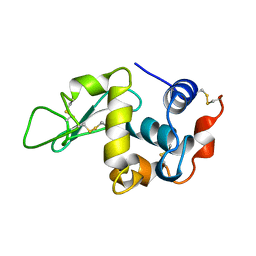 | | THE 1.8 A STRUCTURE OF GROUND CONTROL GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | 分子名称: | PROTEIN (LYSOZYME) | | 著者 | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C. | | 登録日 | 1998-09-24 | | 公開日 | 1998-09-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
255L
 
 | | HYDROLASE | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | 登録日 | 1997-11-10 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1H0Q
 
 | | NMR solution structure of a fully modified locked nucleic acid (LNA) hybridized to RNA | | 分子名称: | 5-D(*(LKC)P*(TLN)P*(LCG)P*(LCA)P*(TLN)P*(LCA)P* (TLN)P*(LCG)P*(LCC))-3, 5-R(*GP*CP*AP*UP*AP*UP*CP*AP*G)-3 | | 著者 | Rasmussen, J, Petersen, M, Nielsen, K.E, Kumar, R, Wengel, J, Jacobsen, J.P. | | 登録日 | 2002-06-27 | | 公開日 | 2003-07-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Studies of Fully Modified Locked Nucleic Acid (Lna) Hybrids: Solution Structure of an Lna:RNA Hybrid and Characterization of an Lna:RNA Hybrid
Bioconjug.Chem., 15, 2004
|
|
1M5L
 
 | |
1C4O
 
 | | CRYSTAL STRUCTURE OF THE DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB FROM THERMUS THERMOPHILUS | | 分子名称: | DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB, SULFATE ION, octyl beta-D-glucopyranoside | | 著者 | Machius, M, Henry, L, Palnitkar, M, Deisenhofer, J. | | 登録日 | 1999-09-14 | | 公開日 | 2000-07-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BL0
 
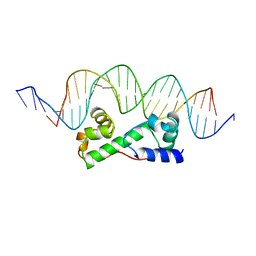 | | MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN (MARA)/DNA COMPLEX | | 分子名称: | DNA (5'-D(*CP*CP*GP*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP* CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP* TP*C)-3'), PROTEIN (MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN) | | 著者 | Davies, S, Rhee, R.G, Martin, J.L, Rosner, D.R. | | 登録日 | 1998-07-22 | | 公開日 | 1998-09-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1M9B
 
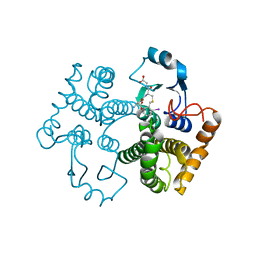 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with gamma-glutamyl[S-(2-iodobenzyl)cysteinyl]glycine | | 分子名称: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-Transferase 26 kDa | | 著者 | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | 登録日 | 2002-07-28 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M99
 
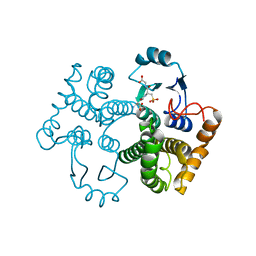 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with glutathione sulfonic acid | | 分子名称: | GLUTATHIONE SULFONIC ACID, Glutathione S-Transferase 26kDa | | 著者 | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | 登録日 | 2002-07-28 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9G
 
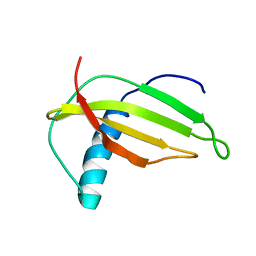 | | Solution structure of G16A-MNEI, a structural mutant of single chain monellin MNEI | | 分子名称: | Monellin chain B and Monellin chain A | | 著者 | Spadaccini, R, Trabucco, F, Saviano, G, Picone, D, Crescenzi, O, Tancredi, T, Temussi, P.A. | | 登録日 | 2002-07-29 | | 公開日 | 2003-06-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Mechanism of Interaction of Sweet Proteins with the T1R2-T1R3 Receptor: Evidence from the Solution Structure of G16A-MNEI
J.MOL.BIOL., 328, 2003
|
|
1MDC
 
 | |
1C1D
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NADH AND L-PHENYLALANINE | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | 著者 | Vanhooke, J.L, Thoden, J.B. | | 登録日 | 1999-07-21 | | 公開日 | 2000-08-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
1C1S
 
 | | RECRUITING ZINC TO MEDIATE POTENT, SPECIFIC INHIBITION OF SERINE PROTEASES | | 分子名称: | BIS(5-AMIDINO-BENZIMIDAZOLYL)METHANE, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Katz, B.A, Luong, C. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Recruiting Zn2+ to mediate potent, specific inhibition of serine proteases.
J.Mol.Biol., 292, 1999
|
|
1VYS
 
 | |
222L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1KEV
 
 | | STRUCTURE OF NADP-DEPENDENT ALCOHOL DEHYDROGENASE | | 分子名称: | NADP-DEPENDENT ALCOHOL DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | 著者 | Korkhin, Y, Frolow, F. | | 登録日 | 1996-10-21 | | 公開日 | 1997-10-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystalline alcohol dehydrogenases from the mesophilic bacterium Clostridium beijerinckii and the thermophilic bacterium Thermoanaerobium brockii: preparation, characterization and molecular symmetry.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
253L
 
 | | LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | 登録日 | 1997-11-10 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
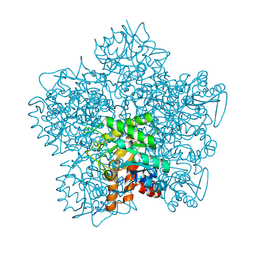
1T34
 
 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | 著者 | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | 登録日 | 2004-04-23 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
1KLO
 
 | |
1TD1
 
 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with acetate | | 分子名称: | ACETATE ION, purine-nucleoside phosphorylase | | 著者 | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | 登録日 | 2004-05-21 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1HFB
 
 | |
1YHS
 
 | | Crystal structure of Pim-1 bound to staurosporine | | 分子名称: | Proto-oncogene serine/threonine-protein kinase Pim-1, STAUROSPORINE | | 著者 | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | 登録日 | 2005-01-10 | | 公開日 | 2005-01-25 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1CLI
 
 | | X-RAY CRYSTAL STRUCTURE OF AMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE (PURM), FROM THE E. COLI PURINE BIOSYNTHETIC PATHWAY, AT 2.5 A RESOLUTION | | 分子名称: | PROTEIN (PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE), SULFATE ION | | 著者 | Li, C, Kappock, T.J, Stubbe, J, Weaver, T.M, Ealick, S.E. | | 登録日 | 1999-04-28 | | 公開日 | 1999-10-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
Structure Fold.Des., 7, 1999
|
|
