6J6G
 
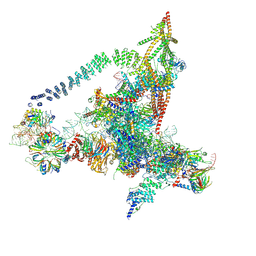 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6BK8
 
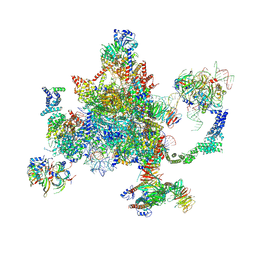 | | S. cerevisiae spliceosomal post-catalytic P complex | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | 著者 | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | 登録日 | 2017-11-07 | | 公開日 | 2018-02-21 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
5ZWM
 
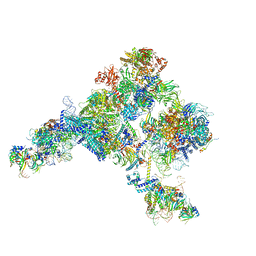 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.4~4.6 angstrom (tri-snRNP and U2 snRNP Part) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, ... | | 著者 | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2018-05-16 | | 公開日 | 2018-08-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5GMK
 
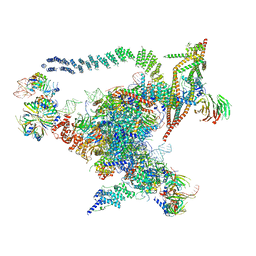 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | 分子名称: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-07-14 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
5Y88
 
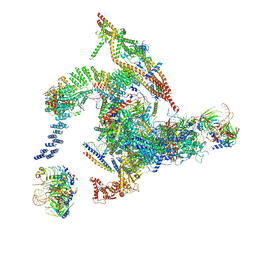 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-08-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5YLZ
 
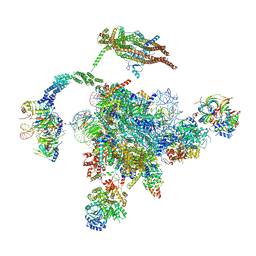 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-10-20 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6EXN
 
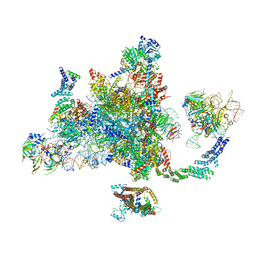 | | Post-catalytic P complex spliceosome with 3' splice site docked | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | 著者 | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | 登録日 | 2017-11-08 | | 公開日 | 2018-01-17 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
6J6Q
 
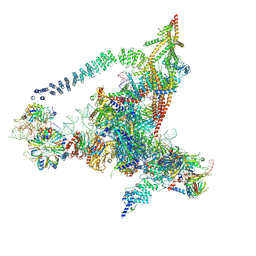 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5LJ3
 
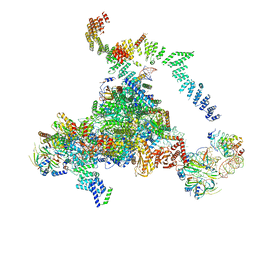 | | Structure of the core of the yeast spliceosome immediately after branching | | 分子名称: | CEF1, CLF1, CWC15, ... | | 著者 | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-03 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | 分子名称: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | 著者 | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-31 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5ZWO
 
 | | Cryo-EM structure of the yeast B complex at average resolution of 3.9 angstrom | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | 著者 | Bai, R, Wan, R, Yan, C, Shi, Y. | | 登録日 | 2018-05-16 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
6G90
 
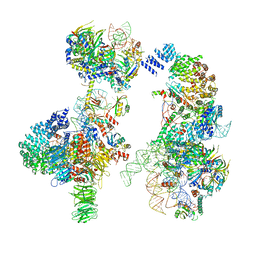 | | Prespliceosome structure provides insight into spliceosome assembly and regulation (map A2) | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing factor 39, ... | | 著者 | Plaschka, C, Lin, P.-C, Charenton, C, Nagai, K. | | 登録日 | 2018-04-10 | | 公開日 | 2018-08-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Prespliceosome structure provides insights into spliceosome assembly and regulation.
Nature, 559, 2018
|
|
5WSG
 
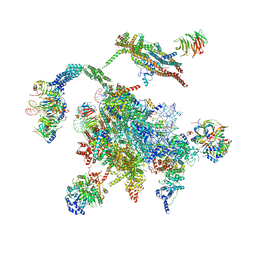 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | 分子名称: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | 著者 | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-12-07 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | 分子名称: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | 著者 | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | 登録日 | 2016-12-19 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
7OQE
 
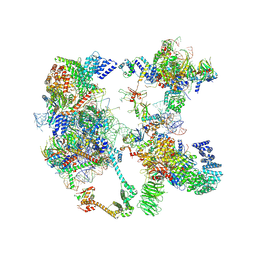 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | 著者 | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | 登録日 | 2021-06-03 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
5NRL
 
 | | Structure of a pre-catalytic spliceosome. | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | 著者 | Plaschka, C, Lin, P.-C, Nagai, K. | | 登録日 | 2017-04-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Structure of a pre-catalytic spliceosome.
Nature, 546, 2017
|
|
7OQB
 
 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | 分子名称: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | 著者 | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | 登録日 | 2021-06-03 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
