3G7E
 
 | |
4ZKI
 
 | |
3G75
 
 | |
3G7B
 
 | |
6TCK
 
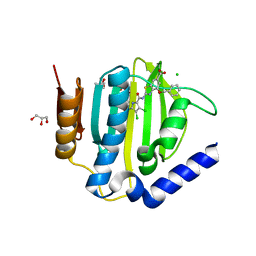 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with ULD-2 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | 著者 | Welin, M, Kimbung, R, Focht, D. | | 登録日 | 2019-11-06 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Rational design of balanced dual-targeting antibiotics with limited resistance.
Plos Biol., 18, 2020
|
|
3H80
 
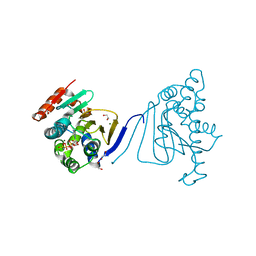 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 | | 分子名称: | 1,2-ETHANEDIOL, Heat shock protein 83-1, MAGNESIUM ION, ... | | 著者 | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2009-04-28 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
To be Published
|
|
5CTW
 
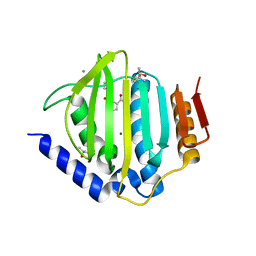 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTX
 
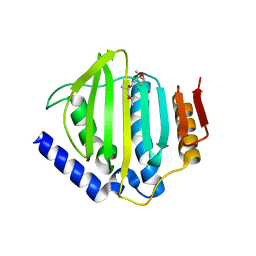 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-21 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5MMP
 
 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)-isoquinolin-3-yl]-urea | | 分子名称: | 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)isoquinolin-3-yl]urea, DNA gyrase subunit B | | 著者 | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMN
 
 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | 分子名称: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | 著者 | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMO
 
 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with [3-(3-ethyl-ureido)-5-(pyridin-4-yl)-isoquinolin-8-yl-methyl]-carbamic acid prop-2-ynyl ester | | 分子名称: | DNA gyrase subunit B, PHOSPHATE ION, prop-2-ynyl ~{N}-[[3-(ethylcarbamoylamino)-5-pyridin-4-yl-isoquinolin-8-yl]methyl]carbamate | | 著者 | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | 登録日 | 2016-12-12 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
1BGQ
 
 | |
1BXD
 
 | | NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ | | 分子名称: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (OSMOLARITY SENSOR PROTEIN (ENVZ)) | | 著者 | Tanaka, T, Saha, S.K, Tomomori, C, Ishima, R, Liu, D, Tong, K.I, Park, H, Dutta, R, Qin, L, Swindells, M.B, Yamazaki, T, Ono, A.M, Kainosho, M, Inouye, M, Ikura, M. | | 登録日 | 1998-10-02 | | 公開日 | 1999-10-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ.
Nature, 396, 1998
|
|
1AJ6
 
 | |
5IDM
 
 | | Bifunctional histidine kinase CckA (domain, CA) in complex with c-di-GMP and AMPPNP/Mg2+ | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Cell cycle histidine kinase CckA, MAGNESIUM ION, ... | | 著者 | Dubey, B.N, Schirmer, T. | | 登録日 | 2016-02-24 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking.
Sci Adv, 2, 2016
|
|
8BN6
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | 著者 | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | 登録日 | 2022-11-12 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
1I5C
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA | | 著者 | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | 登録日 | 2001-02-26 | | 公開日 | 2001-08-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5B
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPNP AND MANGANESE | | 分子名称: | ACETATE ION, CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, ... | | 著者 | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | 登録日 | 2001-02-26 | | 公開日 | 2001-08-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I59
 
 | | STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ADPNP AND MAGENSIUM | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA, ... | | 著者 | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | 登録日 | 2001-02-26 | | 公開日 | 2001-08-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1ID0
 
 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE BOND CONFORMATION OF PHOQ KINASE DOMAIN | | 分子名称: | MAGNESIUM ION, PHOQ HISTIDINE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Marina, A, Mott, C, Auyzenberg, A, Waldburger, C.D, Hendrickson, W.A. | | 登録日 | 2001-04-02 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism.
J.Biol.Chem., 276, 2001
|
|
1A4H
 
 | |
1AM1
 
 | |
1AH8
 
 | |
