5EZK
 
 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | 登録日 | 2015-11-26 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (8.5 Å) | | 主引用文献 | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
6VZ5
 
 | | Escherichia coli transcription-translation complex D3 (TTC-D3) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZ3
 
 | | Escherichia coli transcription-translation complex D2 (TTC-D2) containing mRNA with a 27 nt long spacer | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5MS0
 
 | |
6VYZ
 
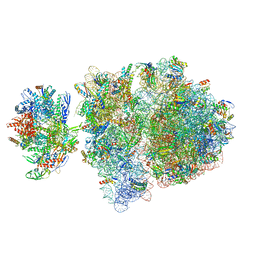 | | Escherichia coli transcription-translation complex C6 (TTC-C6) containing mRNA with a 21 nt long spacer, NusA, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (9.9 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYY
 
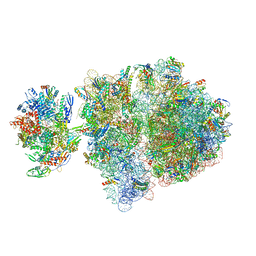 | | Escherichia coli transcription-translation complex C5 (TTC-C5) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (9.9 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYX
 
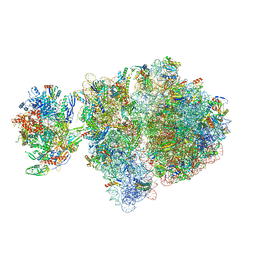 | | Escherichia coli transcription-translation complex C4 (TTC-C4) containing mRNA with a 21 nt long spacer, transcription factor NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (9.9 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZ2
 
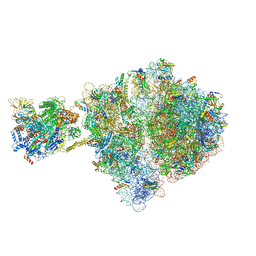 | | Escherichia coli transcription-translation complex D1 (TTC-D1) containing mRNA with a 27 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
3LU0
 
 | | Molecular model of Escherichia coli core RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Darst, S.A. | | 登録日 | 2010-02-16 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.2 Å) | | 主引用文献 | Complete structural model of Escherichia coli RNA polymerase from a hybrid approach.
Plos Biol., 8, 2010
|
|
6N4C
 
 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | 分子名称: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | 登録日 | 2018-11-19 | | 公開日 | 2020-05-27 | | 実験手法 | ELECTRON MICROSCOPY (17 Å) | | 主引用文献 | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | 著者 | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | 登録日 | 2009-08-01 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (19.799999 Å) | | 主引用文献 | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
