6PBS
 
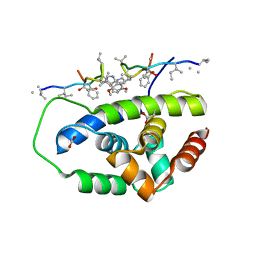 | | Structure of ClpC1-NTD in complex with Ecumicin | | 分子名称: | ACETATE ION, ATP-dependent Clp protease ATP-binding subunit ClpC1, ecumicin | | 著者 | Abad-Zapatero, C, Wolf, N.M. | | 登録日 | 2019-06-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6UCR
 
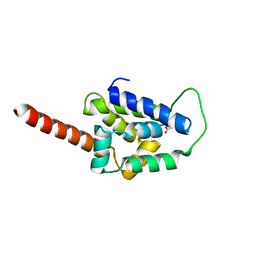 | | Structure of ClpC1-NTD L92S L96P | | 分子名称: | ACETATE ION, Negative regulator of genetic competence ClpC/mecB | | 著者 | Abad-Zapatero, C, Wolf, N.M. | | 登録日 | 2019-09-17 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBA
 
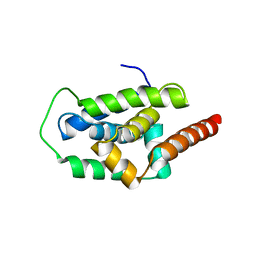 | | Structure of ClpC1-NTD | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpC1 | | 著者 | Abad-Zapatero, C, Wolf, N.M. | | 登録日 | 2019-06-13 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBQ
 
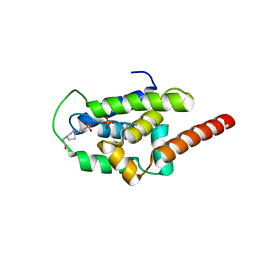 | | Structure of ClpC1-NTD | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp protease ATP-binding subunit ClpC1, PHOSPHATE ION | | 著者 | Abad-Zapatero, C, Wolf, N.M. | | 登録日 | 2019-06-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
