1NLK
 
 | |
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | 分子名称: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | 登録日 | 2003-01-07 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NLN
 
 | | CRYSTAL STRUCTURE OF HUMAN ADENOVIRUS 2 PROTEINASE WITH ITS 11 AMINO ACID COFACTOR AT 1.6 ANGSTROM RESOLUTION | | 分子名称: | ACETIC ACID, Adenain, PVIC | | 著者 | McGrath, W.J, Ding, J, Sweet, R.M, Mangel, W.F. | | 登録日 | 2003-01-07 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold
Biochim.Biophys.Acta, 1648, 2003
|
|
1NLO
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | C-SRC, NL1 (MN7-MN2-MN1-PLPPLP) | | 著者 | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | 登録日 | 1996-08-04 | | 公開日 | 1997-01-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1NLP
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | C-SRC, NL2 (MN8-MN1-PLPPLP) | | 著者 | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | 登録日 | 1996-08-04 | | 公開日 | 1997-01-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1NLQ
 
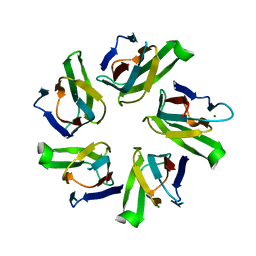 | | The crystal structure of Drosophila NLP-core provides insight into pentamer formation and histone binding | | 分子名称: | MAGNESIUM ION, Nucleoplasmin-like protein | | 著者 | Namboodiri, V.M.H, Dutta, S, Akey, I.V, Head, J.F, Akey, C.W. | | 登録日 | 2003-01-07 | | 公開日 | 2003-03-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of Drosophila NLP-core Provides Insight into Pentamer Formation and Histone Binding
Structure, 11, 2003
|
|
1NLR
 
 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | 分子名称: | ENDO-1,4-BETA-GLUCANASE | | 著者 | Sulzenbacher, G, Dupont, C, Davies, G.J. | | 登録日 | 1997-10-27 | | 公開日 | 1998-11-25 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
1NLS
 
 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | 分子名称: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | 著者 | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | 登録日 | 1997-01-28 | | 公開日 | 1997-11-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.94 Å) | | 主引用文献 | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1NLT
 
 | | The crystal structure of Hsp40 Ydj1 | | 分子名称: | Mitochondrial protein import protein MAS5, Seven residue peptide, ZINC ION | | 著者 | Li, J, Sha, B. | | 登録日 | 2003-01-07 | | 公開日 | 2004-01-13 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate.
Structure, 11, 2003
|
|
1NLU
 
 | | Pseudomonas sedolisin (serine-carboxyl proteinase) complexed with two molecules of pseudo-iodotyrostatin | | 分子名称: | CALCIUM ION, PSEUDO-IODOTYROSTATIN, SEDOLISIN | | 著者 | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Glodfarb, N.E, Dunn, B.M, Oda, K. | | 登録日 | 2003-01-07 | | 公開日 | 2004-01-20 | | 最終更新日 | 2012-12-12 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Two inhibitor molecules bound in the active site of Pseudomonas sedolisin: a model for the bi-product complex following cleavage of a peptide substrate.
Biochem.Biophys.Res.Commun., 314, 2004
|
|
1NLV
 
 | | Crystal Structure Of Dictyostelium Discoideum Actin Complexed With Ca ATP And Human Gelsolin Segment 1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | 著者 | Vorobiev, S.M, Welti, S, Condeelis, J, Almo, S.C. | | 登録日 | 2003-01-07 | | 公開日 | 2003-01-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of Non-Vertebrate Actin: Implications For The ATP Hydrolytic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NLW
 
 | | Crystal structure of Mad-Max recognizing DNA | | 分子名称: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | 著者 | Nair, S.K, Burley, S.K. | | 登録日 | 2003-01-07 | | 公開日 | 2003-02-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1NLX
 
 | | Crystal Structure of PHL P 6, A Major Timothy Grass Pollen Allergen Co-Crystallized with Zinc | | 分子名称: | ARSENIC, Pollen allergen Phl p 6, ZINC ION | | 著者 | Fedorov, A.A, Ball, T, Fedorov, E.V, Vrtala, S, Valenta, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-01-07 | | 公開日 | 2003-01-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure oh Phl p 6, a major timothy grass pollen allergen co-crystallized with Zinc
To be Published
|
|
1NLY
 
 | | Crystal structure of the traffic ATPase of the Helicobacter pylori type IV secretion system in complex with ATPgammaS | | 分子名称: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | 登録日 | 2003-01-08 | | 公開日 | 2003-05-06 | | 最終更新日 | 2012-03-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
1NLZ
 
 | | Crystal structure of unliganded traffic ATPase of the type IV secretion system of helicobacter pylori | | 分子名称: | virB11 homolog | | 著者 | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | 登録日 | 2003-01-08 | | 公開日 | 2003-05-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
1NM0
 
 | | Proteus mirabilis catalase in complex with formiate | | 分子名称: | Catalase, FORMIC ACID, GLYCEROL, ... | | 著者 | Andreoletti, P, Pernoud, A, Gouet, P, Jouve, H.M. | | 登録日 | 2003-01-08 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural studies of Proteus mirabilis catalase in its ground state, oxidized state and in complex with formic acid.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NM1
 
 | | Crystal Structure of D. Dicsoideum Actin Complexed With Gelsolin Segment 1 and Mg ATP at 1.8 A Resolution | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | 著者 | Vorobiev, S.M, Welti, S, Condeelis, J, Almo, S.C. | | 登録日 | 2003-01-08 | | 公開日 | 2003-01-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of Non-Vertebrate Actin: Implications For The ATP Hydrolytic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NM2
 
 | | Malonyl-CoA:ACP Transacylase | | 分子名称: | ACETIC ACID, NICKEL (II) ION, malonyl CoA:acyl carrier protein malonyltransferase | | 著者 | Keatinge-Clay, A.T, Shelat, A.A, Savage, D.F, Tsai, S, Miercke, L.J.W, O'Connell III, J.D, Khosla, C, Stroud, R.M. | | 登録日 | 2003-01-08 | | 公開日 | 2003-01-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Catalysis, Specificity, and ACP Docking Site of
Streptomyces coelicolor Malonyl-CoA:ACP Transacylase
Structure, 11, 2003
|
|
1NM3
 
 | | Crystal structure of Heamophilus influenza hybrid-Prx5 | | 分子名称: | Protein HI0572, SULFATE ION | | 著者 | Kim, S.J, Woo, J.R, Hwang, Y.S, Jeong, D.G, Shin, D.H, Kim, K.H, Ryu, S.E. | | 登録日 | 2003-01-08 | | 公開日 | 2003-03-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Tetrameric Structure of Haemophilus influenza Hybrid Prx5 Reveals Interactions between Electron Donor and Acceptor Proteins.
J.Biol.Chem., 278, 2003
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | 分子名称: | Copper resistance protein C | | 著者 | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-01-09 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NM5
 
 | | R. rubrum transhydrogenase (dI.Q132N)2(dIII)1 asymmetric complex | | 分子名称: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | 著者 | Van Boxel, G.I, Quirk, P.G, Cotton, N.P, White, S.A, Jackson, J.B. | | 登録日 | 2003-01-09 | | 公開日 | 2004-01-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Glutamine 132 in the NAD(H)-binding component of proton-translocating transhydrogenase tethers the nucleotides before hydride transfer.
Biochemistry, 42, 2003
|
|
1NM6
 
 | | thrombin in complex with selective macrocyclic inhibitor at 1.8A | | 分子名称: | (11S)-11-BENZYL-6-CHLORO-1,2,10,11,12,13,14,15,16,17,18,19-DODECAHYDRO-5,9-METHANO-2,5,8,10,13,17-BENZOHEXAAZACYCLOHENICOSINE-3,24-DIONE, Hirudin, thrombin | | 著者 | Nantermet, P.G, Barrow, J.C, Newton, C.L, Pellicore, J.M, Young, M, Lewis, S.D, Lucas, B.J, Krueger, J.A, McMasters, D.R, Yan, Y, Kuo, L.C, Vacca, J.P, Selnick, H.G. | | 登録日 | 2003-01-09 | | 公開日 | 2003-09-02 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design and synthesis of potent and selective macrocyclic thrombin inhibitors
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NM7
 
 | | Solution structure of the ScPex13p SH3 domain | | 分子名称: | Peroxisomal Membrane Protein PAS20 | | 著者 | Pires, J.R, Hong, X, Brockmann, C, Volkmer-Engert, R, Schneider-Mergener, J, Oschkinat, H, Erdmann, R. | | 登録日 | 2003-01-09 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The ScPex13p SH3 Domain Exposes Two Distinct Binding Sites for Pex5p and Pex14p
J.Mol.Biol., 326, 2003
|
|
1NM8
 
 | | Structure of Human Carnitine Acetyltransferase: Molecular Basis for Fatty Acyl Transfer | | 分子名称: | Carnitine O-acetyltransferase | | 著者 | Wu, D, Govindasamy, L, Lian, W, Gu, Y, Kukar, T, Agbandje-McKenna, M, McKenna, R. | | 登録日 | 2003-01-09 | | 公開日 | 2003-03-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Human Carnitine Acetyltransferase. Molecular Basis for Fatty Acyl Transfer
J.Biol.Chem., 278, 2003
|
|
1NM9
 
 | | Crystal structure of recombinant human salivary amylase mutant W58A | | 分子名称: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, Alpha-amylase, ... | | 著者 | Ramasubbu, N, Ragunath, C, Mishra, P.J, Thomas, L.M. | | 登録日 | 2003-01-09 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Human salivary alpha-amylase Trp58 situated at subsite -2 is critical for enzyme activity.
Eur.J.Biochem., 271, 2004
|
|
