5HHA
 
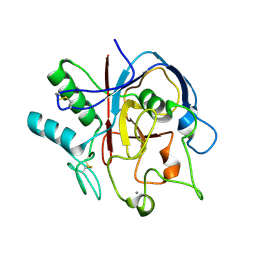 | | Structure of PvdO from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, PvdO | | Authors: | Bai, G, Yuan, Z, Shang, G, Xia, H, Gu, L. | | Deposit date: | 2016-01-10 | | Release date: | 2017-01-18 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Chromophore maturation protein from Pseudomonas aeruginosa
To Be Published
|
|
5AOH
 
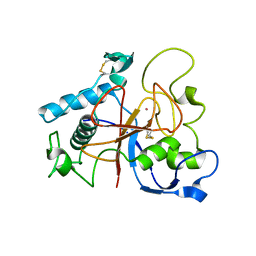 | | Crystal Structure of CarF | | Descriptor: | POTASSIUM ION, Spore coat protein CotH | | Authors: | Tichy, E.M, Hardwick, S.W, Luisi, B.F, C Salmond, G.P. | | Deposit date: | 2015-09-10 | | Release date: | 2017-01-25 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 angstrom resolution crystal structure of the carbapenem intrinsic resistance protein CarF.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7SPQ
 
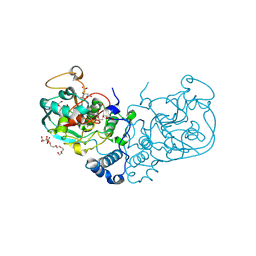 | |
8ARU
 
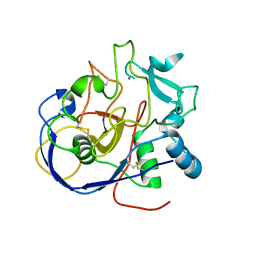 | |
6S07
 
 | | Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen. | | Descriptor: | Abz-ALA-THR-THR-PRO-LEU-CYS-GLY-PRO-SER-ARG-ALA-SER-ILE-LEU-SER-GLY-ARG, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Leisinger, F, Miarzlou, D.A, Seebeck, F.P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Chem Sci, 10, 2019
|
|
2Y3C
 
 | | Treponema denticola variable protein 1 | | Descriptor: | ACETATE ION, CALCIUM ION, SODIUM ION, ... | | Authors: | Le Coq, J, Ghosh, P. | | Deposit date: | 2010-12-20 | | Release date: | 2011-08-31 | | Last modified: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conservation of the C-Type Lectin Fold for Massive Sequence Variation in a Treponema Diversity-Generating Retroelement.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MUJ
 
 | | Formylglycine generating enzyme bound to copper | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Lafrance-Vanasse, J, Appel, M.J, Tsai, C.-L, Bertozzi, C, Tainer, J.A. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Formylglycine-generating enzyme binds substrate directly at a mononuclear Cu(I) center to initiate O2activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2Q17
 
 | | Formylglycine Generating Enzyme from Streptomyces coelicolor | | Descriptor: | CALCIUM ION, formylglycine generating enzyme | | Authors: | Carlson, B.L, Ballister, E.R, Skordalakes, E, King, D.S, Breidenbach, M.A, Gilmore, S.A, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2007-05-23 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function and structure of a prokaryotic formylglycine-generating enzyme.
J.Biol.Chem., 283, 2008
|
|
2HI8
 
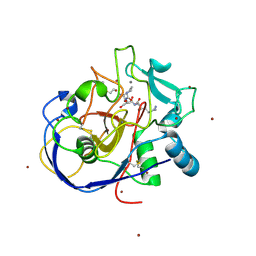 | | human formylglycine generating enzyme, C336S mutant, bromide co-crystallization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Rudolph, M.G, Roeser, D. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5NYY
 
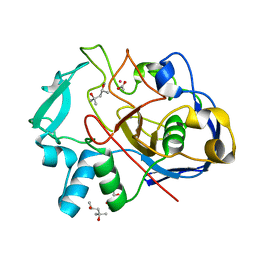 | | Formylglycine generating enzyme from T. curvata in complex with Cd(II) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Meury, M, Knop, M, Seebeck, F.P. | | Deposit date: | 2017-05-12 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Basis for Copper-Oxygen Mediated C-H Bond Activation by the Formylglycine-Generating Enzyme.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NXL
 
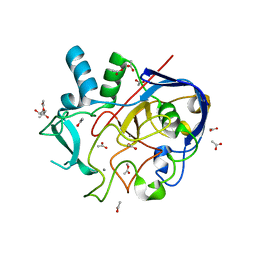 | | Formylglycine generating enzyme from T. curvata in complex with Ag(I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Meury, M, Knop, M, Seebeck, F.P. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural Basis for Copper-Oxygen Mediated C-H Bond Activation by the Formylglycine-Generating Enzyme.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7MSY
 
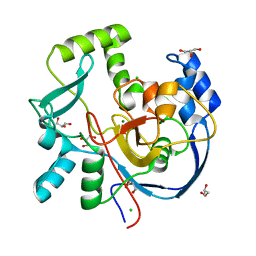 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalU17, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ML6
 
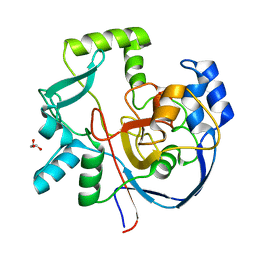 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
2HIB
 
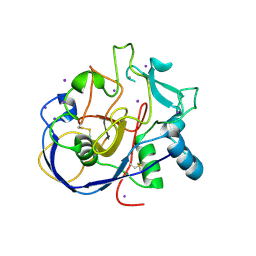 | | human formylglycine generating enzyme, C336S mutant, iodide co-crystallization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Rudolph, M.G, Roeser, D. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5SSX
 
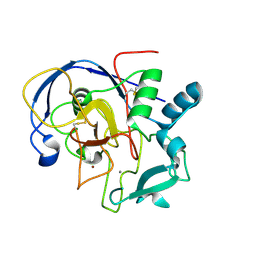 | |
5SSY
 
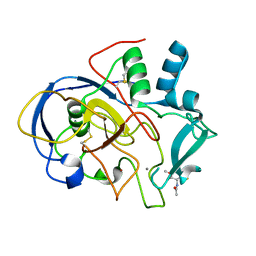 | |
5SSZ
 
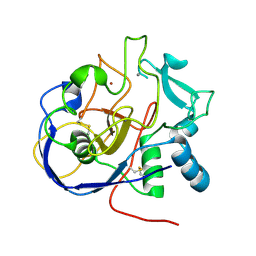 | |
8VIG
 
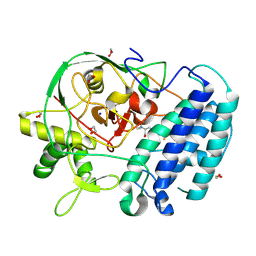 | | EgtB-IV from Geminocystis sp. isolate SKYG4, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with hercynine | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N,N-trimethyl-histidine, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 2024
|
|
8VII
 
 | | EgtB-IV from Crocosphaera subtropica, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with cysteine and hercynine | | Descriptor: | 1,2-ETHANEDIOL, CYSTEINE, MANGANESE (II) ION, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 2024
|
|
8VIK
 
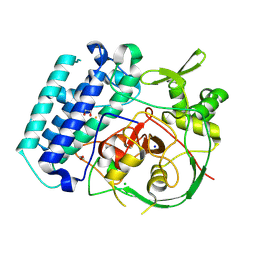 | | EgtB-IV from Crocosphaera subtropica, a type IV sulfoxide synthase involved in ergothioneine biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 2024
|
|
8VIL
 
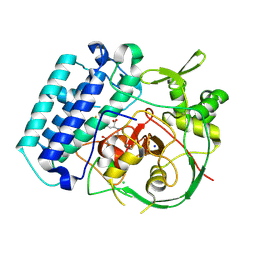 | | EgtB-IV from Crocosphaera subtropica, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with N,N-dimethyl-histidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 2024
|
|
8VIH
 
 | | EgtB-IV from Crocosphaera subtropica, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 2024
|
|
6VQP
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, CalU17 His-Tagged protein, GLYCEROL, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina
To Be Published
|
|
6XTP
 
 | |
6XTQ
 
 | |
