3U7B
 
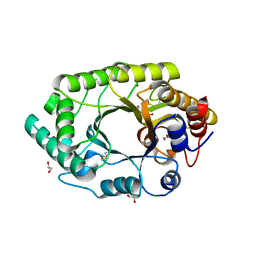 | | A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of a GH10 xylanase from Fusarium oxysporum reveals the presence of an extended loop on top of the catalytic cleft.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8BBI
 
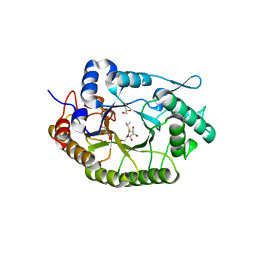 | |
6WQW
 
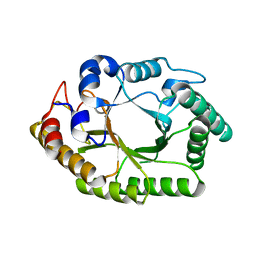 | | Thermobacillus composti GH10 xylanase | | Descriptor: | Beta-xylanase, beta-D-xylopyranose | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Transformation of xylan into value-added biocommodities using Thermobacillus composti GH10 xylanase.
Carbohydr Polym, 247, 2020
|
|
5XC0
 
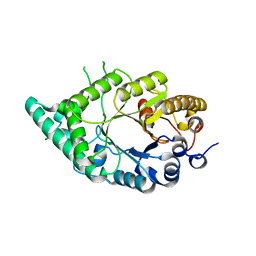 | |
4W8L
 
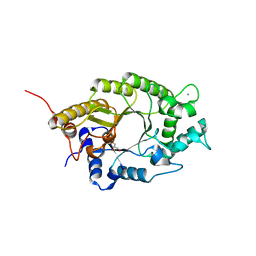 | | Structure of GH10 from Paenibacillus barcinonensis | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase C, GLYCEROL | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-08-25 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring Multimodularity in Plant Cell Wall Deconstruction: STRUCTURAL AND FUNCTIONAL ANALYSIS OF Xyn10C CONTAINING THE CBM22-1-CBM22-2 TANDEM.
J.Biol.Chem., 290, 2015
|
|
4F8X
 
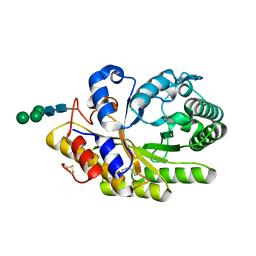 | | Penicillium canescens endo-1,4-beta-xylanase XylE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-1,4-beta-xylanase, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Polyakov, K.M, Trofimov, A.A, Fedorova, T.V, Koroleva, O.V, Maisuradze, I.G, Chulkin, A.M, Vavilova, E.A, Benevolenskii, S.V. | | Deposit date: | 2012-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: |
|
|
6LPS
 
 | |
7NL2
 
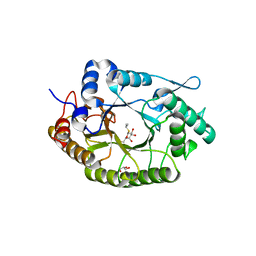 | | Structure of Xyn11 from Pseudothermotoga thermarum | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-xylanase, GLYCEROL | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phylogenetic, functional and structural characterization of a GH10 xylanase active at extreme conditions of temperature and alkalinity
Comput Struct Biotechnol J, 19, 2021
|
|
2XYL
 
 | | CELLULOMONAS FIMI XYLANASE/CELLULASE COMPLEXED WITH 2-DEOXY-2-FLUORO-XYLOBIOSE | | Descriptor: | BETA-1,4-GLYCANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Monem, V, Birsan, C, Warren, R.A.J, Withers, S.G, Rose, D.R. | | Deposit date: | 1997-11-20 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the cellulose/xylan specificity of the beta-1,4-glycanase cex from Cellulomonas fimi through crystallography and mutation.
Biochemistry, 37, 1998
|
|
8B73
 
 | |
8HKG
 
 | |
5M0K
 
 | |
6D5C
 
 | |
2Q8X
 
 | | The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus | | Descriptor: | GLYCEROL, SODIUM ION, intra-cellular xylanase | | Authors: | Solomon, V, Teplitsky, A, Gilboa, R, Zolotnitsky, G, Golan, G, Reiland, V, Moryles, S, Shoham, Y, Shoham, G. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4XUY
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger | | Descriptor: | GLYCEROL, Probable endo-1,4-beta-xylanase C, SULFATE ION | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0027 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger
To Be Published
|
|
8XY0
 
 | | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase A | | Authors: | Nakamura, T, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
1W32
 
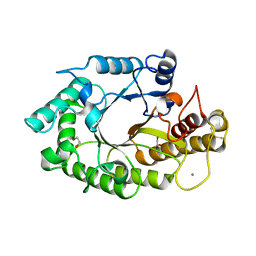 | | The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Andrews, S, Taylor, E.J, Pell, G.N, Vincent, F, Ducros, V.M.A, Davies, G.J, Lakey, J.H, Gilbert, H.J. | | Deposit date: | 2004-07-12 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
8Y1M
 
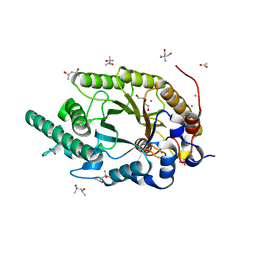 | | Xylanase R from Bacillus sp. TAR-1 complexed with xylobiose. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Nakamura, T, Kuwata, K, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-25 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
1W2V
 
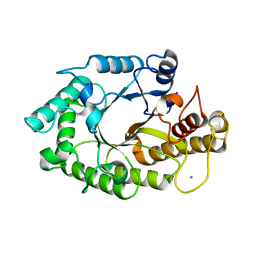 | | The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Andrews, S, Taylor, E.J, Pell, G.N, Vincent, F, Ducros, V.M.A, Davies, G.J, Lakey, J.H, Glbert, H.J. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
4XV0
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei | | Descriptor: | Beta-xylanase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9697 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei
To Be Published
|
|
4XX6
 
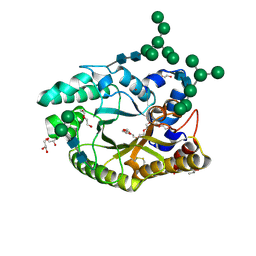 | | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Stogios, P.J, Nocek, B, Xu, X, Cui, H, Lowden, M, Savchenko, A. | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum.
To Be Published
|
|
2HIS
 
 | | CELLULOMONAS FIMI XYLANASE/CELLULASE DOUBLE MUTANT E127A/H205N WITH COVALENT CELLOBIOSE | | Descriptor: | CELLULOMONAS FIMI FAMILY 10 BETA-1,4-GLYCANASE, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Notenboom, V, Birsan, C, Nitz, M, Rose, D.R, Warren, R.A.J, Wither, S.G. | | Deposit date: | 1998-02-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into transition state stabilization of the beta-1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants.
Nat.Struct.Biol., 5, 1998
|
|
4E4P
 
 | |
3RO8
 
 | |
5AY7
 
 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | xylanase | | Authors: | Zheng, Y, Li, Y, Liu, W, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
