1UHV
 
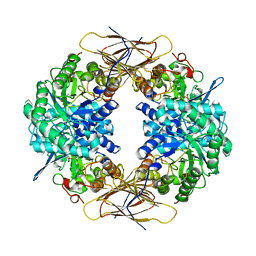 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1UHW
 
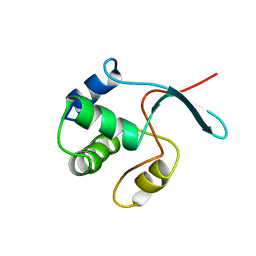 | | Solution structure of the DEP domain of mouse pleckstrin | | Descriptor: | Pleckstrin | | Authors: | Inoue, K, Yoshida, M, Hatta, R, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-11 | | Release date: | 2004-01-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DEP domain of mouse pleckstrin
To be Published
|
|
1UHX
 
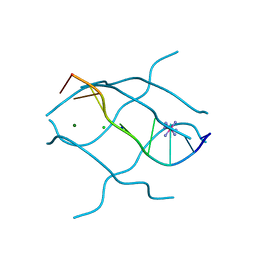 | | Crystal structure of d(GCGAGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UHY
 
 | | Crystal structure of d(GCGATAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*TP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UHZ
 
 | | Solution structure of dsRNA binding domain in Staufen homolog 2 | | Descriptor: | staufen (RNA binding protein) homolog 2 | | Authors: | He, F, Muto, Y, Obayashi, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Koboyashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dsRNA binding domain in Staufen homolog 2
To be Published
|
|
1UI0
 
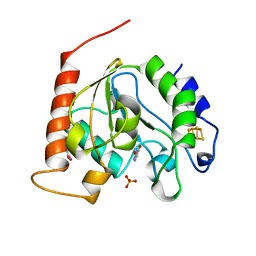 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UI1
 
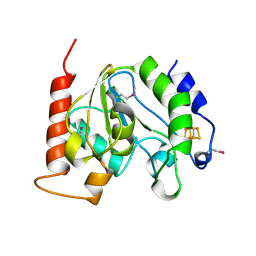 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
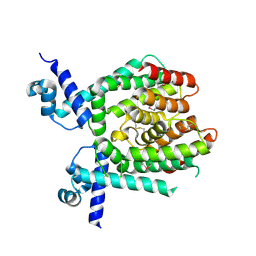
1UI5
 
 | |
1UI6
 
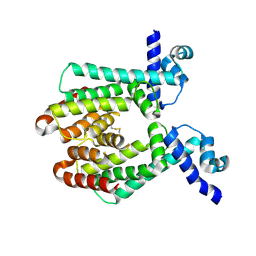 | |
1UI7
 
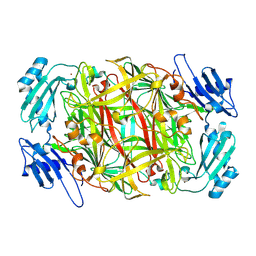 | | Site-directed mutagenesis of His433 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
1UI8
 
 | | Site-directed mutagenesis of His592 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
1UI9
 
 | | Crystal analysis of chorismate mutase from thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, chorismate mutase | | Authors: | Inagaki, E, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of chorismate mutase from thermus thermophilus
To be Published
|
|
1UIA
 
 | |
1UIB
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UIC
 
 | |
1UID
 
 | |
1UIE
 
 | |
1UIF
 
 | |
1UIG
 
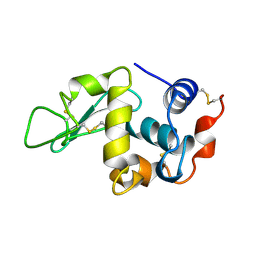 | |
1UIH
 
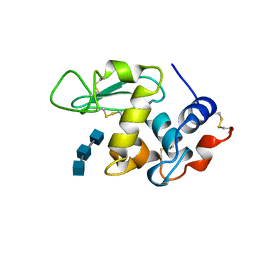 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UII
 
 | |
1UIJ
 
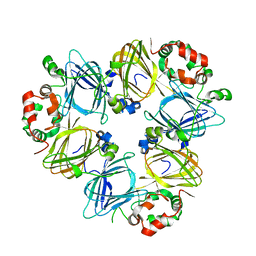 | | Crystal Structure Of Soybean beta-Conglycinin Beta Homotrimer (I122M/K124W) | | Descriptor: | beta subunit of beta conglycinin | | Authors: | Maruyama, N, Maruyama, Y, Tsuruki, T, Okuda, E, Yoshikawa, M, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Creation of soybean beta-conglycinin beta with strong phagocytosis-stimulating activity
BIOCHIM.BIOPHYS.ACTA, 1648, 2003
|
|
1UIK
 
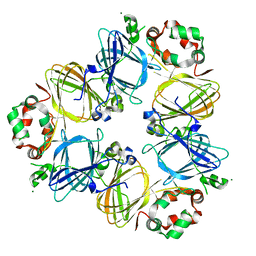 | | Crystal structure of soybean beta-conglycinin alpha prime homotrimer | | Descriptor: | MAGNESIUM ION, alpha prime subunit of beta-conglycinin | | Authors: | Maruyama, Y, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the core region of the soybean beta-conglycinin alpha' subunit.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UIL
 
 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | Descriptor: | Double-stranded RNA-binding motif | | Authors: | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
1UIM
 
 | |
