1UH5
 
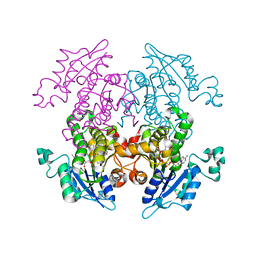 | | Crystal Structure of Enoyl-ACP Reductase with Triclosan at 2.2angstroms | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, enoyl-ACP reductase | | Authors: | Swarnamukhi, P.L, Kapoor, M, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2003-06-24 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the variation in triclosan affinity to enoyl reductases.
J.Mol.Biol., 343, 2004
|
|
1UH6
 
 | | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06 | | Descriptor: | ubiquitin-like 5 | | Authors: | Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-25 | | Release date: | 2003-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06
To be Published
|
|
1UH7
 
 | |
1UH8
 
 | |
1UH9
 
 | |
1UHA
 
 | | Crystal Structure of Pokeweed Lectin-D2 | | Descriptor: | CALCIUM ION, lectin-D2 | | Authors: | Fujii, T, Hayashida, M, Hamasu, M, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-06-27 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of two lectins from the roots of pokeweed (Phytolacca americana).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UHB
 
 | | Crystal structure of porcine alpha trypsin bound with auto catalyticaly produced native peptide at 2.15 A resolution | | Descriptor: | 9-mer peptide from Trypsin, ACETATE ION, CALCIUM ION, ... | | Authors: | Pattabhi, V, Syed Ibrahim, B, Shamaladevi, N. | | Deposit date: | 2003-06-27 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Trypsin activity reduced by an autocatalytically produced nonapeptide.
J.Biomol.Struct.Dyn., 21, 2004
|
|
1UHC
 
 | | Solution Structure of RSGI RUH-002, a SH3 Domain of KIAA1010 protein [Homo sapiens] | | Descriptor: | KIAA1010 protein | | Authors: | Abe, T, Hirota, H, Kobayashi, N, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-28 | | Release date: | 2003-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-002, a SH3 Domain of KIAA1010 protein [Homo sapiens]
To be Published
|
|
1UHD
 
 | | Crystal structure of aspartate decarboxylase, pyruvoly group bound form | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
1UHE
 
 | | Crystal structure of aspartate decarboxylase, isoaspargine complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, N~2~-(2-AMINO-1-METHYL-2-OXOETHYLIDENE)ASPARAGINATE | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
1UHF
 
 | | Solution Structure of the third SH3 domain of human intersectin 2(KIAA1256) | | Descriptor: | INTERSECTIN 2 | | Authors: | Suzuki, S, Hatanaka, H, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-03 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the third SH3 domain of human intersectin 2(KIAA1256)
To be Published
|
|
1UHG
 
 | | Crystal Structure of S-Ovalbumin At 1.9 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ovalbumin, ... | | Authors: | Yamasaki, M, Takahashi, N, Hirose, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of S-ovalbumin as a Non-loop-inserted Thermostabilized Serpin Form
J.Biol.Chem., 278, 2003
|
|
1UHH
 
 | | Crystal structure of cp-aequorin | | Descriptor: | (8R)-8-(CYCLOPENTYLMETHYL)-2-HYDROPEROXY-2-(4-HYDROXYBENZYL)-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H) -ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHI
 
 | | Crystal structure of i-aequorin | | Descriptor: | (2R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(4-IODOBENZYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHJ
 
 | | Crystal structure of br-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-(4-BROMOBENZYL)-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHK
 
 | | Crystal structure of n-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(2-NAPHTHYLMETHYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHL
 
 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
1UHM
 
 | | Solution structure of the globular domain of linker histone homolog Hho1p from S. cerevisiae | | Descriptor: | Histone H1 | | Authors: | Ono, K, Kusano, O, Shimotakahara, S, Shimizu, M, Yamazaki, T, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-05 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Nucleic Acids Res., 31, 2003
|
|
1UHN
 
 | | The crystal structure of the calcium binding protein AtCBL2 from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, calcineurin B-like protein 2 | | Authors: | Nagae, M, Nozawa, A, Koizumi, N, Sano, H, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2003-07-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Novel Calcium-binding Protein AtCBL2 from Arabidopsis thaliana
J.Biol.Chem., 278, 2003
|
|
1UHO
 
 | | Crystal structure of Human Phosphodiesterase 5 complexed with Vardenafil(Levitra) | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-07-09 | | Release date: | 2004-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
1UHP
 
 | | Solution structure of RSGI RUH-005, a PDZ domain in human cDNA, KIAA1095 | | Descriptor: | hypothetical protein KIAA1095 | | Authors: | Hamada, T, Muto, Y, Hirota, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-005, a PDZ domain in human cDNA, KIAA1095
To be published
|
|
1UHR
 
 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
1UHT
 
 | | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein | | Descriptor: | expressed protein | | Authors: | Tomizawa, T, Inoue, M, Koshiba, S, Hayashi, F, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Shinozaki, K, Seki, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein
To be Published
|
|
1UHU
 
 | | Solution structure of the retroviral Gag MA-like domain of RIKEN cDNA 3110009E22 | | Descriptor: | product of RIKEN cDNA 3110009E22 | | Authors: | Suzuki, S, Hatanaka, H, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the retroviral Gag MA-like domain of RIKEN cDNA 3110009E22
To be Published
|
|
