1BUI
 
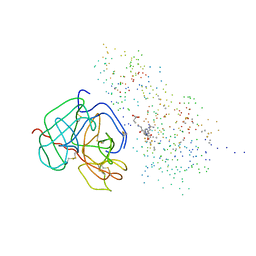 | | Structure of the ternary microplasmin-staphylokinase-microplasmin complex: a proteinase-cofactor-substrate complex in action | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, Plasminogen, Staphylokinase | | Authors: | Parry, M.A.A, Fernandez-Catalan, C, Bergner, A, Huber, R, Hopfner, K, Schlott, B, Guehrs, K, Bode, W. | | Deposit date: | 1998-09-04 | | Release date: | 1999-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The ternary microplasmin-staphylokinase-microplasmin complex is a proteinase-cofactor-substrate complex in action.
Nat.Struct.Biol., 5, 1998
|
|
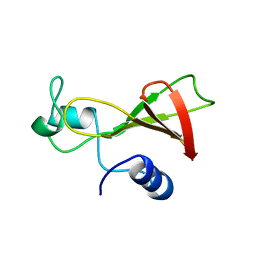
1BUJ
 
 | |
1BUL
 
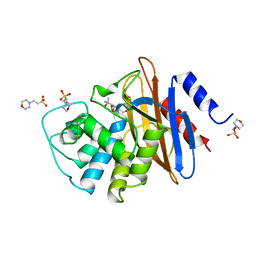 | | 6ALPHA-(HYDROXYPROPYL)PENICILLANATE ACYLATED ON NMC-A BETA-LACTAMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | 2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NMC-A BETA-LACTAMASE | | Authors: | Mourey, L, Swaren, P, Miyashita, K, Bulychev, A, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-04 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Inhibition of the Nmc-A B-Lactamase by a Penicillanic Acid Derivative, and the Structural Bases for the Increase in Substrate Profile of This Antibiotic Resistance Enzyme
J.Am.Chem.Soc., 120, 1998
|
|
1BUN
 
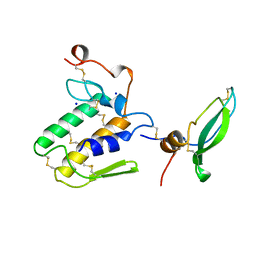 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1BUO
 
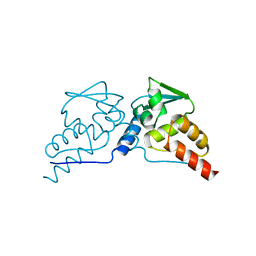 | | BTB DOMAIN FROM PLZF | | Descriptor: | PROTEIN (PROMYELOCYTIC LEUKEMIA ZINC FINGER PROTEIN PLZF) | | Authors: | Ahmad, K.F, Engel, C.K, Prive, G.G. | | Deposit date: | 1998-09-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the BTB domain from PLZF.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BUP
 
 | | T13S MUTANT OF BOVINE 70 KILODALTON HEAT SHOCK PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sousa, M.C, Mckay, D.B. | | Deposit date: | 1998-09-03 | | Release date: | 1998-09-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change.
Biochemistry, 37, 1998
|
|
1BUQ
 
 | | SOLUTION STRUCTURE OF DELTA-5-3-KETOSTEROID ISOMERASE COMPLEXED WITH THE STEROID 19-NORTESTOSTERONE-HEMISUCCINATE | | Descriptor: | PROTEIN (3-KETOSTEROID ISOMERASE-19-NORTESTOSTERONE-HEMISUCCINATE), SUCCINIC ACID MONO-(13-METHYL-3-OXO-2,3,6,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYCLOPENTA[A]PHENANTHREN-17-YL) ESTER | | Authors: | Massiah, M.A, Abeygunawardana, C, Gittis, A.G, Mildvan, A.S. | | Deposit date: | 1998-09-04 | | Release date: | 1999-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Delta 5-3-ketosteroid isomerase complexed with the steroid 19-nortestosterone hemisuccinate.
Biochemistry, 37, 1998
|
|
1BUS
 
 | |
1BUT
 
 | |
1BUU
 
 | |
1BUV
 
 | | CRYSTAL STRUCTURE OF THE MT1-MMP-TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, PROTEIN (MEMBRANE-TYPE MATRIX METALLOPROTEINASE (CDMT1-MMP)), PROTEIN (METALLOPROTEINASE INHIBITOR (TIMP-2)), ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-09-07 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
1BUW
 
 | |
1BUX
 
 | | 3'-PHOSPHORYLATED NUCLEOTIDES BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Xu, Y, Schneider, B, Deville-Bonne, D, Veron, M, Janin, J. | | Deposit date: | 1998-09-07 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3'-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity.
J.Biol.Chem., 273, 1998
|
|
1BUY
 
 | | HUMAN ERYTHROPOIETIN, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN (ERYTHROPOIETIN) | | Authors: | Cheetham, J.C, Smith, D.M, Aoki, K.H, Stevenson, J.L, Hoeffel, T.J, Syed, R.S, Egrie, J, Harvey, T.S. | | Deposit date: | 1998-09-08 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human erythropoietin and a comparison with its receptor bound conformation.
Nat.Struct.Biol., 5, 1998
|
|
1BUZ
 
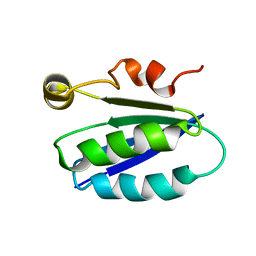 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BV1
 
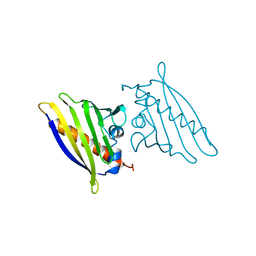 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | BET V 1 | | Authors: | Gajhede, M, Osmark, P, Poulsen, F.M, Ipsen, H, Larson, J.N, Joostvan, R.J, Schou, C, Lowenstein, H, Spangfort, M.D. | | Deposit date: | 1997-07-08 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
1BV2
 
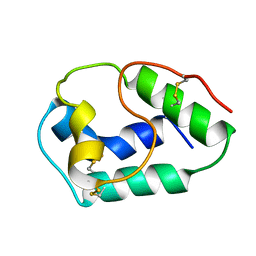 | | LIPID TRANSFER PROTEIN FROM RICE SEEDS, NMR, 14 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Poznanski, J, Sodano, P, Suh, S.W, Lee, J.Y, Ptak, M, Vovelle, F. | | Deposit date: | 1998-09-21 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins.
Eur.J.Biochem., 259, 1999
|
|
1BV3
 
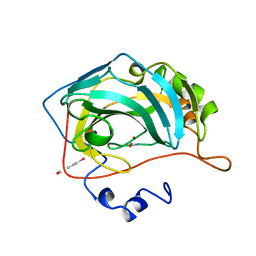 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH UREA | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, PROTEIN (CARBONIC ANHYDRASE II), UREA, ... | | Authors: | Briganti, F, Mangani, S, Scozzafava, A, Vernaglione, G, Supuran, C.T. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Carbonic anhydrase catalyzes cyanamide hydration to urea: is it mimicking the physiological reaction?
J.Biol.Inorg.Chem., 4, 1999
|
|
1BV4
 
 | | APO-MANNOSE-BINDING PROTEIN-C | | Descriptor: | PROTEIN (MANNOSE-BINDING PROTEIN-C) | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1998-09-22 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ca2+-dependent structural changes in C-type mannose-binding proteins.
Biochemistry, 37, 1998
|
|
1BV7
 
 | | COUNTERACTING HIV-1 PROTEASE DRUG RESISTANCE: STRUCTURAL ANALYSIS OF MUTANT PROTEASES COMPLEXED WITH XV638 AND SD146, CYCLIC UREA AMIDES WITH BROAD SPECIFICITIES | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BV8
 
 | |
1BV9
 
 | | HIV-1 PROTEASE (I84V) COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BVA
 
 | | MANGANESE BINDING MUTANT IN CYTOCHROME C PEROXIDASE | | Descriptor: | MANGANESE (II) ION, PROTEIN (CYTOCHROME C PEROXIDASE), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Rational design of a functional metalloenzyme: introduction of a site for manganese binding and oxidation into a heme peroxidase.
Biochemistry, 37, 1998
|
|
1BVB
 
 | | HEME-PACKING MOTIFS REVEALED BY THE CRYSTAL STRUCTURE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C-554, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Iverson, T.M, Arciero, D.M, Hsu, B.T, Logan, M.S.P, Hooper, A.B, Rees, D.C. | | Deposit date: | 1998-09-16 | | Release date: | 1999-05-18 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea.
Nat.Struct.Biol., 5, 1998
|
|
1BVC
 
 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM D) AT 118 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA, PHOSPHATE ION | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
