1AET
 
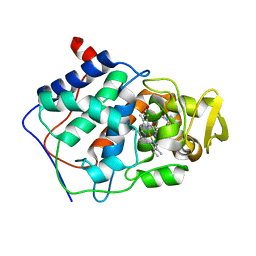 | | VARIATION IN THE STRENGTH OF A CH TO O HYDROGEN BOND IN AN ARTIFICIAL PROTEIN CAVITY (1-METHYLIMIDAZOLE) | | Descriptor: | 1-METHYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1AHX
 
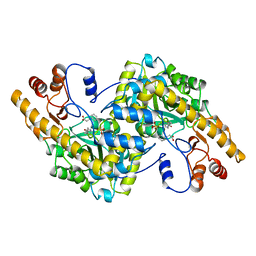 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | ASPARTATE AMINOTRANSFERASE, HYDROCINNAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
1A8Q
 
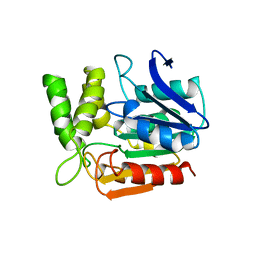 | | BROMOPEROXIDASE A1 | | Descriptor: | BROMOPEROXIDASE A1 | | Authors: | Hofmann, B, Toelzer, S, Pelletier, I, Altenbuchner, J, Van Pee, K.-H, Hecht, H.-J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural investigation of the cofactor-free chloroperoxidases.
J.Mol.Biol., 279, 1998
|
|
1AIB
 
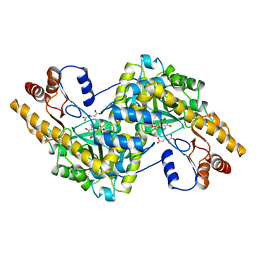 | |
8JMT
 
 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | Descriptor: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | Authors: | Tao, Y, Guo, Q, He, B, Zhong, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
1A82
 
 | | DETHIOBIOTIN SYNTHETASE FROM ESCHERICHIA COLI, COMPLEX WITH SUBSTRATES ATP AND DIAMINOPELARGONIC ACID | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, DETHIOBIOTIN SYNTHETASE, ... | | Authors: | Kaeck, H, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshot of a phosphorylated substrate intermediate by kinetic crystallography.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AEJ
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (1-VINYLIMIDAZOLE) | | Descriptor: | 1-VINYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
7DQA
 
 | | Cryo-EM structure of SARS-CoV2 RBD-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, J, Lan, J, Wang, X.Q, Wang, H.W. | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reduced graphene oxide membrane as supporting film for high-resolution cryo-EM
Biophys Rep, 7, 2022
|
|
8JLX
 
 | | CCHFV envelope protein Gc in complex with Gc13 | | Descriptor: | Glycoprotein C,CCHFV Gc fusion loops, Mouse antibody Gc13 heavy chain, Mouse antibody Gc13 light chain | | Authors: | Chong, T, Cao, S. | | Deposit date: | 2023-06-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
8J7M
 
 | | ion channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, ion channel,Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment) | | Authors: | Chen, H.W, Chen, H.W. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | ion channel
To Be Published
|
|
7DPE
 
 | | RNA methyltransferase METTL4 | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*GP*TP*GP*AP*TP*CP*AP*CP*GP*CP*GP*GP*C)-3'), GLYCEROL, Methyltransferase-like protein 2, ... | | Authors: | Luo, Q, Ma, J.B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structure of RNA m6A methyltransferase METTL4 in complex with DNA at 2.75 Angstroms resolutioon
To Be Published
|
|
1A14
 
 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
7DR1
 
 | | Structure of Wild-type PSI monomer2 from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
8IWM
 
 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | Descriptor: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWE
 
 | | Cryo-EM structure of the SPE-mTAAR9 complex | | Descriptor: | SPERMIDINE, Trace amine-associated receptor 9 | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8ONS
 
 | |
7DR2
 
 | | Structure of GraFix PSI tetramer from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
1AEB
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (3-METHYLTHIAZOLE) | | Descriptor: | 3-METHYLTHIAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
7DR0
 
 | | Structure of Wild-type PSI monomer1 from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
1AEO
 
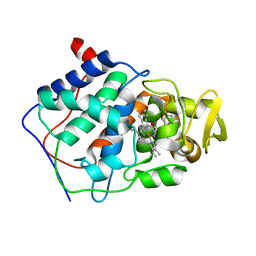 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (2-AMINOPYRIDINE) | | Descriptor: | 2-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
7DRX
 
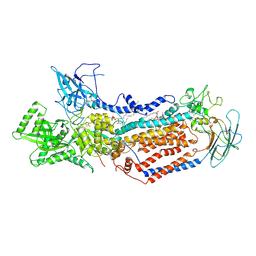 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-30 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
8OX4
 
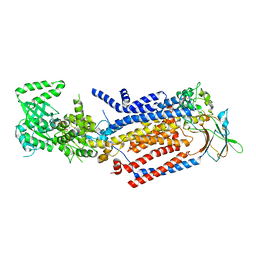 | | Cryo-EM structure of ATP8B1-CDC50A in E1-ATP conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, MAGNESIUM ION, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
1ADO
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT MUSCLE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ALDOLASE, SULFATE ION | | Authors: | Blom, N.S, Sygusch, J. | | Deposit date: | 1996-12-02 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product binding and role of the C-terminal region in class I D-fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 4, 1997
|
|
1AIA
 
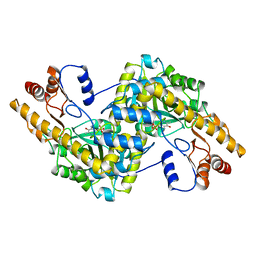 | |
8OX9
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P active conformation with bound PC | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
