8RZC
 
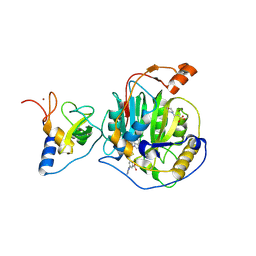 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
6HR4
 
 | |
7Z98
 
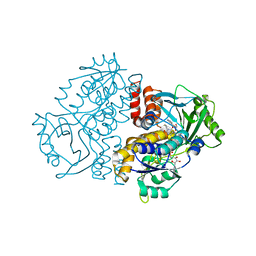 | | Crystal structure of F191M variant Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with methyl phenyl sulfide | | Descriptor: | (methylsulfanyl)benzene, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6ZQQ
 
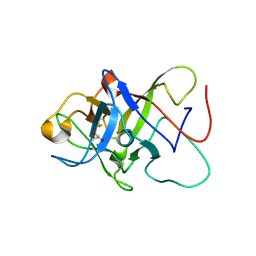 | | Structure of the Pmt3-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT3 isoform 1 | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
8RZD
 
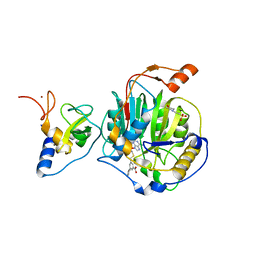 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
6HSA
 
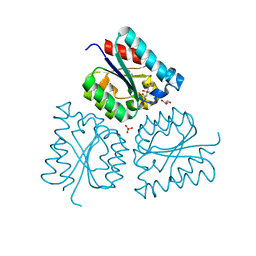 | |
6BG8
 
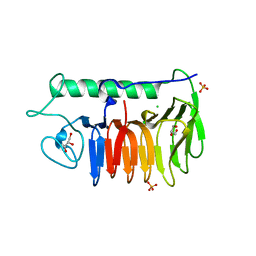 | |
7PJN
 
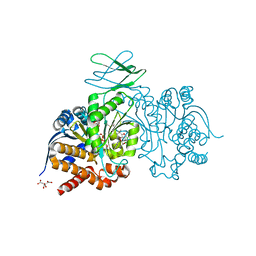 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
6HNL
 
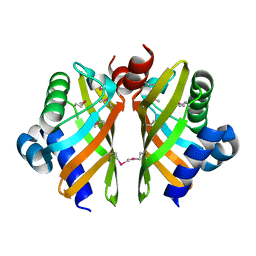 | | Selenomethionine derivative of IdmH 96-104 loop truncation variant | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
7KND
 
 | | C1B domain of Protein kinase C in apo form | | Descriptor: | Protein kinase C delta type, ZINC ION | | Authors: | Katti, S.S, Krieger, I. | | Deposit date: | 2020-11-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7ZCA
 
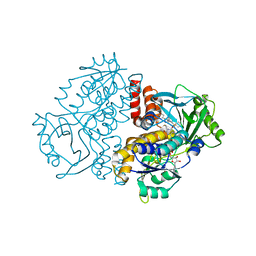 | | Crystal structure of the F191M/F201A variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with benzyl phenyl sulfoxide | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative dehydrogenase/oxygenase subunit (Flavoprotein), ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-26 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6HSN
 
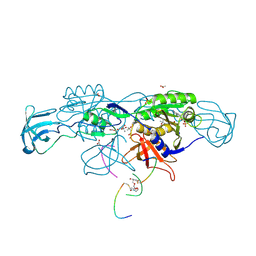 | | Crystal structure of the ternary complex of GephE-ADP-GABA(A) receptor derived peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis(1H-pyrrole-2,5-dione), ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
6B2B
 
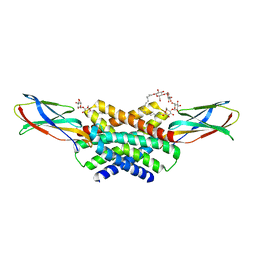 | | Crystal structure of fluoride channel Fluc Ec2 F83M Mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, FLUORIDE ION, Fluoride ion transporter CrcB, ... | | Authors: | Last, N.B, Sun, S, Pham, M.C, Miller, C. | | Deposit date: | 2017-09-19 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular determinants of permeation in a fluoride-specific ion channel.
Elife, 6, 2017
|
|
8R7A
 
 | |
6HTA
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Glutathione transferase Xi 3 C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
7Z6E
 
 | |
7Z99
 
 | |
7PSE
 
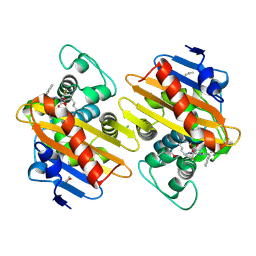 | | Crystal Structure of a Class D Carbapenemase_K73ALY Complexed with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, 1-BUTANOL, Beta-lactamase, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
6ZYI
 
 | |
7PUP
 
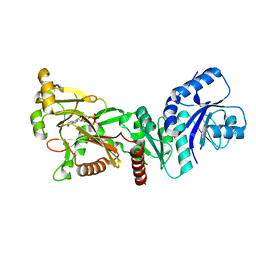 | | INOSITOL 1,3,4-TRISPHOSPHATE 5/6-KINASE 4 from Arabidopsis thaliana (AtITPK4) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol 1,3,4-trisphosphate 5/6-kinase 4 | | Authors: | Whitfield, H, He, S, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversification in the inositol tris/tetrakisphosphate kinase (ITPK) family: crystal structure and enzymology of the outlier AtITPK4.
Biochem.J., 480, 2023
|
|
7Z97
 
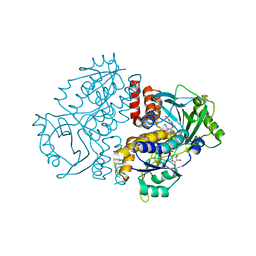 | | Crystal structure of the F191M variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with 6-bromoindole | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6B85
 
 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
8RV5
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 1 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
7YXF
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-(2-piperazin-1-ylethyl)-5-pyridin-4-yl-indole, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-02-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7PSX
 
 | | Structure of HOXB13 bound to hydroxymethylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*5HCP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2021-09-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HOXB13 bound to hydroxymethylated DNA
To Be Published
|
|
