7UR4
 
 | |
8OPC
 
 | |
8OPD
 
 | |
1N21
 
 | | (+)-Bornyl Diphosphate Synthase: Cocrystal with Mg and 3-aza-2,3-dihydrogeranyl diphosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[METHYL-(4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid synthase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6N06
 
 | |
8GYP
 
 | |
8GYT
 
 | | Cryo-EM structure of human Pannexin-3 R36S/F40R variant in pre-open state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pannexin-3 | | Authors: | Cong, Y, Jin, X, Kong, F, Wu, T, Yan, C. | | Deposit date: | 2022-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Inner channel lipids regulated gating mechanism of human pannexins.
To Be Published
|
|
1YMO
 
 | |
5IRX
 
 | | Structure of TRPV1 in complex with DkTx and RTX, determined in lipid nanodisc | | Descriptor: | (2S)-2-(acetyloxy)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}propyl pentanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-14 | | Release date: | 2016-05-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
6N8Z
 
 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6N0G
 
 | |
8OPB
 
 | |
1I45
 
 | | YEAST TRIOSEPHOSPHATE ISOMERASE (MUTANT) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rozovsky, S, Jogl, G, Tong, L, McDermott, A.E. | | Deposit date: | 2001-02-19 | | Release date: | 2001-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Solution-state NMR investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics.
J.Mol.Biol., 310, 2001
|
|
7U51
 
 | |
1I50
 
 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
7U52
 
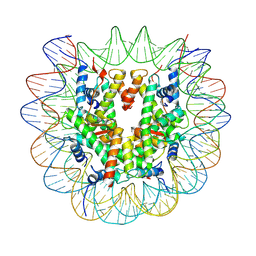 | |
1IA0
 
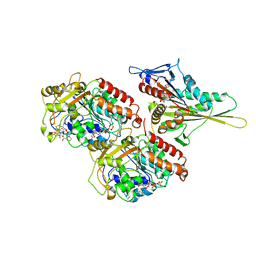 | | KIF1A HEAD-MICROTUBULE COMPLEX STRUCTURE IN ATP-FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-22 | | Release date: | 2002-03-22 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Switch-based Mechanism of Kinesin Motors
Nature, 411, 2001
|
|
4AOR
 
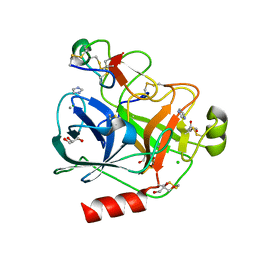 | | Cationic trypsin in complex with the Spinacia oleracea trypsin inhibitor III (SOTI-III) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6MZY
 
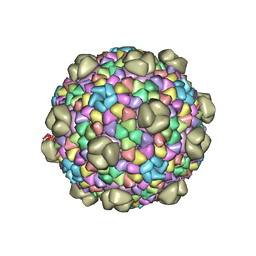 | |
7ATY
 
 | | Solution NMR structure of the SH3 domain of human Caskin1 | | Descriptor: | Caskin-1 | | Authors: | Toke, O, Koprivanacz, K, Radnai, L, Mero, B, Juhasz, T, Liliom, K, Buday, L. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the SH3 Domain of Human Caskin1 Validates the Lack of a Typical Peptide Binding Groove and Supports a Role in Lipid Mediator Binding.
Cells, 10, 2021
|
|
7B2F
 
 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1PPQ
 
 | | NMR structure of 16th module of Immune Adherence Receptor, Cr1 (Cd35) | | Descriptor: | Complement receptor type 1 | | Authors: | O'Leary, J.M, Bromek, K, Black, G.M, Uhrinova, S, Schmitz, C, Krych, M, Atkinson, J.P, Uhrin, D, Barlow, P.N. | | Deposit date: | 2003-06-17 | | Release date: | 2004-05-04 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces.
Protein Sci., 13, 2004
|
|
7U53
 
 | |
6SL1
 
 | | Structure of the open conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SKZ
 
 | | Structure of the closed conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
