1ZA6
 
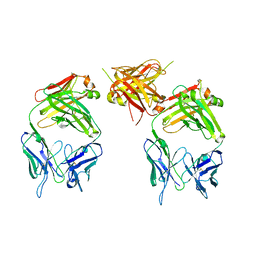 | | The structure of an antitumor CH2-domain-deleted humanized antibody | | Descriptor: | IGG Heavy chain, IGG Light chain | | Authors: | Larson, S.B, Day, J.S, Glaser, S, Braslawsky, G, McPherson, A. | | Deposit date: | 2005-04-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of an Antitumor C(H)2-domain-deleted Humanized Antibody.
J.Mol.Biol., 348, 2005
|
|
1Z8D
 
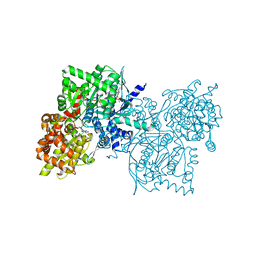 | | Crystal Structure of Human Muscle Glycogen Phosphorylase a with AMP and Glucose | | Descriptor: | ADENINE, ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Lukacs, C.M, Oikonomakos, N.G, Crowther, R.L, Hong, L.N, Kammlott, R.U, Levin, W, Li, S, Liu, C.M, Lucas-McGady, D, Pietranico, S, Reik, L. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: An intermediate conformation with T-state and R-state features.
Proteins, 63, 2006
|
|
1UVY
 
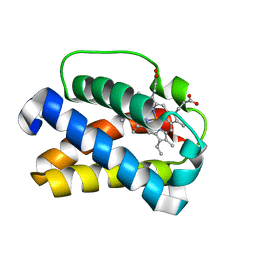 | | HEME-LIGAND TUNNELING IN GROUP I TRUNCATED HEMOGLOBINS | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, XENON | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heme-Ligand Tunneling in Group I Truncated Hemoglobins
J.Biol.Chem., 279, 2004
|
|
1UOC
 
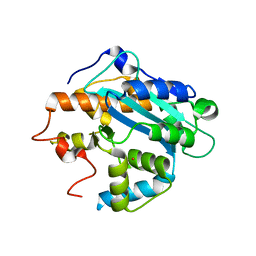 | | X-ray structure of the RNase domain of the yeast Pop2 protein | | Descriptor: | CALCIUM ION, POP2, XENON | | Authors: | Thore, S, Mauxion, F, Seraphin, B, Suck, D. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure and Activity of the Yeast Pop2 Protein: A Nuclease Subunit of the Mrna Deadenylase Complex
Embo Rep., 4, 2003
|
|
1ZBD
 
 | |
1V3E
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with ZANAMAVIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1ZI6
 
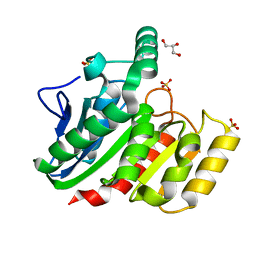 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZIN
 
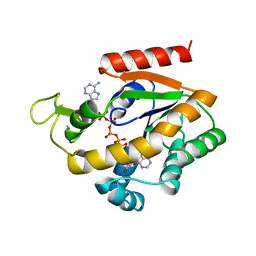 | | ADENYLATE KINASE WITH BOUND AP5A | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1996-06-07 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
1V5H
 
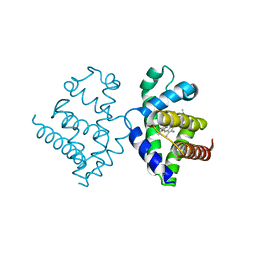 | | Crystal Structure of Human Cytoglobin (Ferric Form) | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Makino, M, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of human cytoglobin for ligand binding.
J.Mol.Biol., 339, 2004
|
|
1ZDJ
 
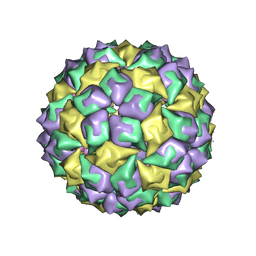 | | STRUCTURE OF BACTERIOPHAGE COAT PROTEIN-LOOP RNA COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*C)-3') | | Authors: | Grahn, E, Stonehouse, N, Valegard, K, Vandenworm, S, Liljas, L. | | Deposit date: | 1997-12-18 | | Release date: | 1998-07-08 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of RNA hairpins in complexes with recombinant MS2 capsids: implications for binding requirements.
RNA, 5, 1999
|
|
2AXH
 
 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
2AY9
 
 | |
1VC0
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Imidazole and Sr2+ solution | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VCD
 
 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase Ndx1 | | Descriptor: | GLYCEROL, Ndx1, SULFATE ION | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
1ZL3
 
 | | Coupling of active site motions and RNA binding | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*CP*GP*GP*UP*(FLO) UP*CP*GP*AP*UP*CP*CP*CP*GP*UP*UP*GP*C)-3', SULFATE ION, tRNA pseudouridine synthase B | | Authors: | Hoang, C, Hamilton, C.S, Mueller, E.G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain
Protein Sci., 14, 2005
|
|
1UZB
 
 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
2BAM
 
 | |
2AZK
 
 | | Crystal structure for the mutant W136E of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
1VBB
 
 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R80633 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE BUTYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1ZHY
 
 | | Structure of yeast oxysterol binding protein Osh4 in complex with cholesterol | | Descriptor: | CHOLESTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
2B1C
 
 | |
2B1K
 
 | | Crystal structure of E. coli CcmG protein | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Ouyang, N, Gao, Y.G, Hu, H.Y, Xia, Z.X. | | Deposit date: | 2005-09-15 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region
Proteins, 65, 2006
|
|
1UN9
 
 | | Crystal structure of the dihydroxyacetone kinase from C. freundii in complex with AMP-PNP and Mg2+ | | Descriptor: | DIHYDROXYACETONE, DIHYDROXYACETONE KINASE, MAGNESIUM ION, ... | | Authors: | Siebold, C, Arnold, I, Garcia-Alles, L.F, Baumann, U, Erni, B. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Citrobacter Freundii Dihydroxyacetone Kinase Reveals an Eight-Stranded Alpha-Helical Barrel ATP-Binding Domain
J.Biol.Chem., 278, 2003
|
|
1ZNS
 
 | | Crystal structure of N-ColE7/12-bp DNA/Zn complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3', Colicin E7, ZINC ION | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
1UNS
 
 | | IDENTIFICATION OF A SECONDARY ZINC-BINDING SITE IN STAPHYLOCOCCAL ENTEROTOXIN C2: IMPLICATIONS FOR SUPERANTIGEN RECOGNITION | | Descriptor: | ENTEROTOXIN TYPE C-2, ZINC ION | | Authors: | Papageorgiou, A.C, Baker, M.D, McLeod, J.D, Goda, S, Sansom, D.M, Tranter, H.S, Acharya, K.R. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Secondary Zinc-Binding Site in Staphylococcal Enterotoxin C2: Implications for Superantigen Recognition
J.Biol.Chem., 279, 2004
|
|
