6OGJ
 
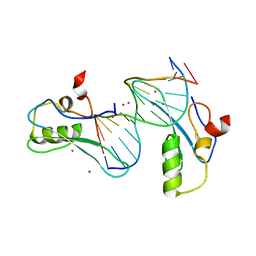 | | MeCP2 MBD in complex with DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*CP*AP*CP*TP*CP*CP*G)-3'), Methyl-CpG-binding protein 2, ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
6U15
 
 | |
4XQK
 
 | |
2LEX
 
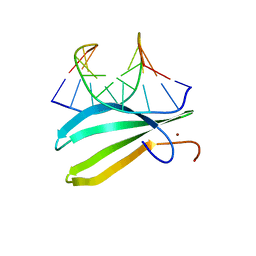 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
1QRV
 
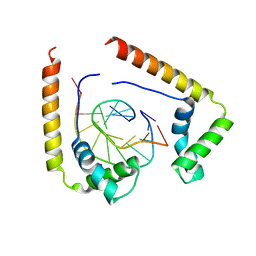 | | CRYSTAL STRUCTURE OF THE COMPLEX OF HMG-D AND DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3'), HIGH MOBILITY GROUP PROTEIN D, SODIUM ION | | Authors: | Murphy IV, F.V, Sweet, R.M, Churchill, M.E.A. | | Deposit date: | 1999-06-15 | | Release date: | 1999-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition.
EMBO J., 18, 1999
|
|
3FLO
 
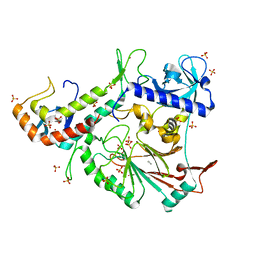 | |
257D
 
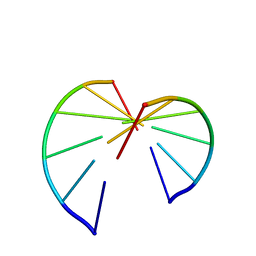 | |
256D
 
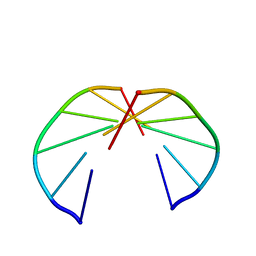 | |
131D
 
 | | THE LOW-TEMPERATURE CRYSTAL STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA REVEALS BINDING OF A SPERMINE MOLECULE IN THE MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SODIUM ION, SPERMINE | | Authors: | Bancroft, D, Williams, L.D, Rich, A, Egli, M. | | Deposit date: | 1993-06-18 | | Release date: | 1993-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove.
Biochemistry, 33, 1994
|
|
145D
 
 | |
4O3N
 
 | | Crystal structure of human dna polymerase eta in ternary complex with native dna and incoming nucleotide (dcp) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
6OZI
 
 | |
220D
 
 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*AP*CP*CP*CP*GP*CP*GP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
1A35
 
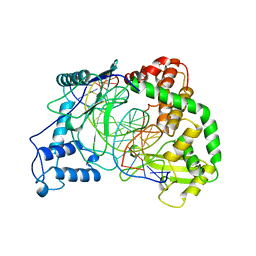 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
8H0L
 
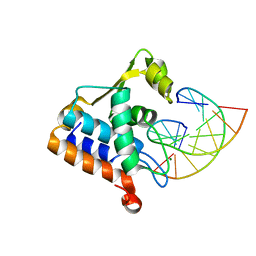 | | Sulfur binding domain of Hga complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*(PST)P*TP*CP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*GP*AP*AP*CP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Liu, G, He, X, Hu, W, Yang, B, Xiao, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a promiscuous DNA sulfur binding domain and application in site-directed RNA base editing.
Nucleic Acids Res., 51, 2023
|
|
4FZX
 
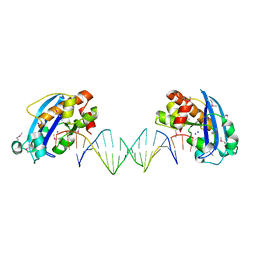 | | Exonuclease X in complex with 3' overhanging duplex DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*TP*CP*CP*AP*CP*AP*AP*TP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, ... | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
5D3G
 
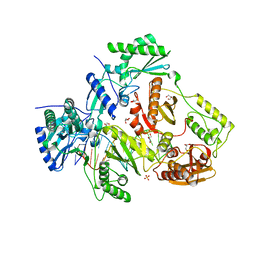 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | Descriptor: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
5NL0
 
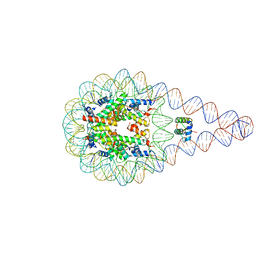 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
2BNA
 
 | |
4FXD
 
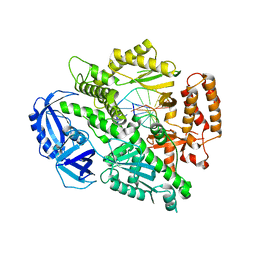 | | Crystal structure of yeast DNA polymerase alpha bound to DNA/RNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*CP*GP*CP*TP*GP*CP*CP*CP*GP*CP*CP*T)-3'), DNA polymerase alpha catalytic subunit A, RNA (5'-R(*AP*GP*GP*CP*GP*GP*GP*CP*AP*G)-3') | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-07-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism for priming DNA synthesis by yeast DNA Polymerase alpha
eLife, 2, 2013
|
|
7CQ4
 
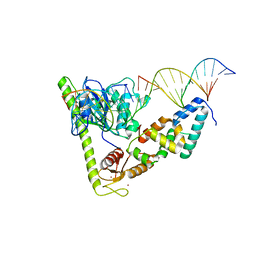 | | Crystal structure of Slx1-Slx4 in complex with 5'flap DNA | | Descriptor: | DNA (27-MER), DNA (5'-D(*AP*GP*GP*AP*CP*AP*TP*CP*TP*TP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*TP*AP*CP*AP*AP*CP*AP*GP*AP*T)-3'), ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
212D
 
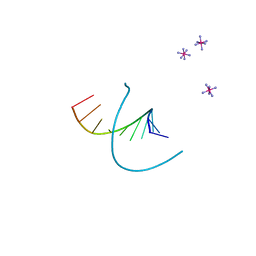 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
8APM
 
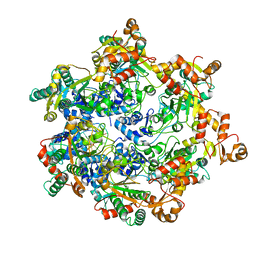 | | Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C1 | | Descriptor: | DNA (5'-D(P*CP*CP*GP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*TP*TP*CP*GP*G)-3'), Primase D5 | | Authors: | Burmeister, W.P, Hutin, S, Ling, W.L, Grimm, C, Schoehn, G. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction.
Viruses, 14, 2022
|
|
5NW5
 
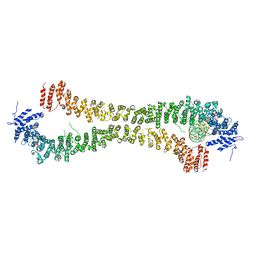 | | Crystal structure of the Rif1 N-terminal domain (RIF1-NTD) from Saccharomyces cerevisiae in complex with DNA | | Descriptor: | DNA (30-MER), DNA (60-MER), Telomere length regulator protein RIF1 | | Authors: | Bunker, R.D, Reinert, J.K, Shi, T, Thoma, N.H. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.502 Å) | | Cite: | Rif1 maintains telomeres and mediates DNA repair by encasing DNA ends.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3V6D
 
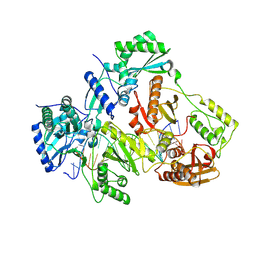 | | Crystal structure of HIV-1 reverse transcriptase (RT) cross-linked with AZT-terminated DNA | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7048 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
