6VGQ
 
 | | ClpP1P2 complex from M. tuberculosis with GLF-CMK bound to ClpP1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, Z-Gly-leu-phe-CH2Cl | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FG4
 
 | | HRAS R97L Crystal Form 1 | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Fetics, S, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
3TMM
 
 | | TFAM imposes a U-turn on mitochondrial DNA | | Descriptor: | DNA (28-MER), Transcription factor A, mitochondrial | | Authors: | Ngo, H.B, Kaiser, J.T, Chan, D.C. | | Deposit date: | 2011-08-31 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3OOI
 
 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
3U3U
 
 | | Crystal structure of the tablysin-15-leukotriene E4 complex | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, CITRIC ACID, PRASEODYMIUM ION, ... | | Authors: | Andersen, J.F. | | Deposit date: | 2011-10-06 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain.
J.Biol.Chem., 287, 2012
|
|
6VGK
 
 | | ClpP1P2 complex from M. tuberculosis | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3S0X
 
 | |
8FG3
 
 | | HRAS R97L Crystal Form 2 | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Fetics, S, Johnson, C.W, Mattos, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
2X4U
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
4QHN
 
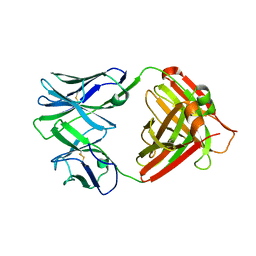 | | I2 (unbound) from CH103 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5KP4
 
 | | Crystal Structure of Ketosteroid Isomerase from Pseudomonas putida (pKSI) bound to 19-nortestosterone | | Descriptor: | (8~{R},9~{S},10~{R},13~{S},14~{S},17~{S})-13-methyl-17-oxidanyl-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1~{H}-cyclop enta[a]phenanthren-3-one, Steroid Delta-isomerase | | Authors: | Wu, Y, Boxer, S.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | A Critical Test of the Electrostatic Contribution to Catalysis with Noncanonical Amino Acids in Ketosteroid Isomerase.
J.Am.Chem.Soc., 138, 2016
|
|
3SG8
 
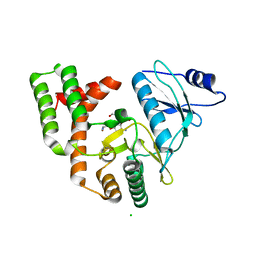 | |
4CW0
 
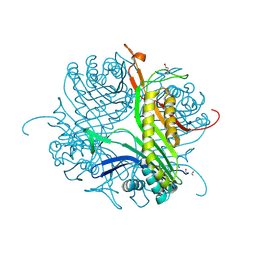 | |
4QHM
 
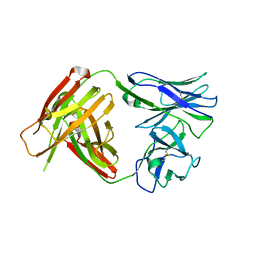 | | I3.1 (unbound) from CH103 Lineage | | Descriptor: | I2 light chain, I3 heavy chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6FG1
 
 | |
6N7Q
 
 | |
2LUQ
 
 | |
3OVO
 
 | |
6N95
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-CoA | | Descriptor: | (2R)-sulfonatepropionyl-CoA, (2S)-sulfonatepropionyl-CoA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
3SG9
 
 | |
3PEP
 
 | |
3SI5
 
 | | Kinetochore-BUBR1 kinase complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Protein CASC5 | | Authors: | Blundell, T.L, Chirgadze, D.Y, Bolanos-Garcia, V.M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Blinkin-BUBR1 Complex Reveals an Interaction Crucial for Kinetochore-Mitotic Checkpoint Regulation via an Unanticipated Binding Site.
Structure, 19, 2011
|
|
3SKF
 
 | |
3SOS
 
 | | Benzothiazinone inhibitor in complex with FXIa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, Coagulation factor XI, ... | | Authors: | Fradera, X, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-30 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | High-resolution crystal structures of factor XIa coagulation factor in complex with nonbasic high-affinity synthetic inhibitors.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3PEC
 
 | |
