3TLF
 
 | |
3H9K
 
 | | Structures of Thymidylate Synthase R163K with Substrates and Inhibitors Show Subunit Asymmetry | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Gibson, L.M, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5FLC
 
 | | Architecture of human mTOR Complex 1 - 5.9 Angstrom reconstruction | | Descriptor: | FKBP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, REGULATORY-ASSOCIATED PROTEIN OF MTOR, ... | | Authors: | Aylett, C.H.S, Sauer, E, Imseng, S, Boehringer, D, Hall, M.N, Ban, N, Maier, T. | | Deposit date: | 2015-10-23 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Architecture of Human Mtor Complex 1
Science, 351, 2016
|
|
4MV6
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Phosphonoacetamide | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOACETAMIDE | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MVO
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
1KGZ
 
 | | Crystal Structure Analysis of the Anthranilate Phosphoribosyltransferase from Erwinia carotovora (current name, Pectobacterium carotovorum) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MANGANESE (II) ION, ... | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-28 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
4MK2
 
 | | 3-(5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzonitrile bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzonitrile, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
3H6D
 
 | | Structure of the mycobacterium tuberculosis DUTPase D28N mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Nagy, G, Takacs, E, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-04-23 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
4MP0
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5FNU
 
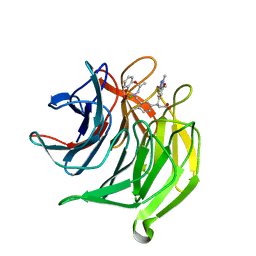 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
2YUS
 
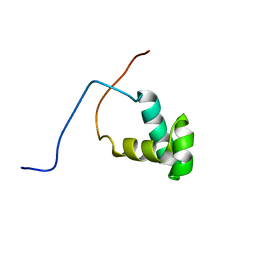 | | Solution structure of the SANT domain of human SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 | | Authors: | Tochio, N, Koshiba, S, Satio, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SANT domain of human SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1
To be Published
|
|
2N4L
 
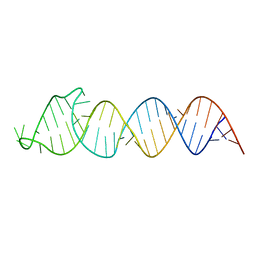 | | Solution Structure of the HIV-1 Intron Splicing Silencer and its Interactions with the UP1 Domain of hnRNP A1 | | Descriptor: | RNA (53-MER) | | Authors: | Tolbert, B.S, Jain, N, Morgan, C.E, Rife, B.D, Salemi, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Intron Splicing Silencer and Its Interactions with the UP1 Domain of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) A1.
J.Biol.Chem., 291, 2016
|
|
4MVU
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MW8
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.256 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MUM
 
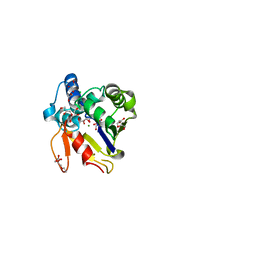 | |
2Z16
 
 | | Crystal structure of Matrix protein 1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1) | | Descriptor: | Matrix protein 1 | | Authors: | Saijo, S, Kishishita, S, Uchikubo-Kamo, T, Terada, T, Shirouzu, M, Ito, H, Ito, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Matrix protein 1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1)
To be Published
|
|
4MV9
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Biotin carboxylase | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
8CCD
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CI7
 
 | |
7A8Y
 
 | |
8CCB
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((1R)-1-(((2S)-1-(2-(5-chlorothiophen-2-yl)-2-cyclohexylacetyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)-2-cyclohexyl-ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
7ABD
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-768 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,4-dimethyl-1~{H}-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-768
To be published
|
|
8CCE
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)butanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)butanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCC
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)propanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)propanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCF
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)pent-4-enoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)pent-4-enoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
