8PVF
 
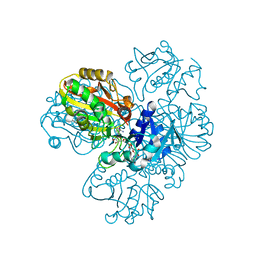 | | Structure of GAPDH determined by cryoEM at 100 keV | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7B2F
 
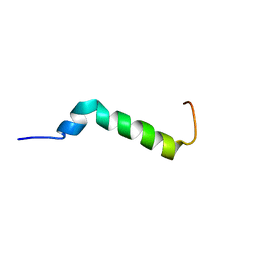 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B2B
 
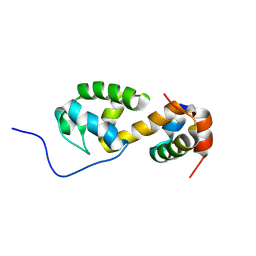 | | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | | Descriptor: | Amino acid adenylation domain-containing protein, Peptide synthetase PaxA | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7RTX
 
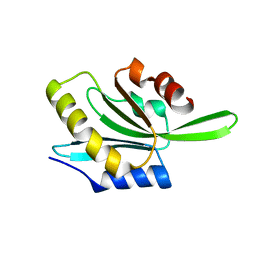 | | Actophorin grown in microgravity | | Descriptor: | Actophorin | | Authors: | Lieberman, R.L, Quirk, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improved resolution crystal structure of Acanthamoeba actophorin reveals structural plasticity not induced by microgravity.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7DEQ
 
 | | Lysozyme-sugar complex in D2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DER
 
 | | Lysozyme alone in H2O | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7BID
 
 | | Crystal structure of v31WRAP-T, a 7-bladed designer protein | | Descriptor: | v31WRAP-T | | Authors: | Laier, I, Mylemans, B, Lee, X.Y, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIF
 
 | | Crystal structure of v22WRAP-T, a 7-bladed designer protein | | Descriptor: | v22WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIG
 
 | | Crystal structure of v13WRAP-T, a 7-bladed designer protein | | Descriptor: | CHLORIDE ION, v13WRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7BIE
 
 | | Crystal structure of nvWrap-T, a 7-bladed symmetric propeller | | Descriptor: | CITRIC ACID, nvWRAP-T | | Authors: | Lee, X.Y, Mylemans, B, Laier, I, Voet, A.R.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the designer protein WRAP-T and its permutants.
Sci Rep, 11, 2021
|
|
7WNW
 
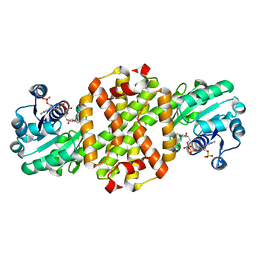 | |
7WNN
 
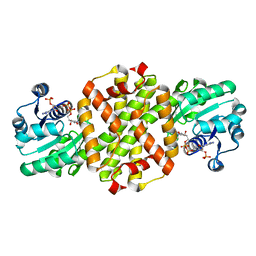 | |
7W5C
 
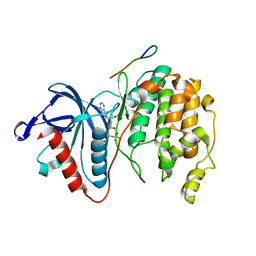 | | Crystal structure of Mitogen Activated Protein Kinase 4 (MPK4) from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 4, Mitogen-activated protein kinase kinase 1, ... | | Authors: | Arold, S.T, Hameed, U.F.S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Essential role of the CD docking motif of MPK4 in plant immunity, growth, and development.
New Phytol., 239, 2023
|
|
7Z3H
 
 | | Crystal structure of Iba57 from Chaetomium thermophilum | | Descriptor: | CITRIC ACID, Transferase-like protein | | Authors: | Altegoer, F, Mrusek, D, Bange, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The iron-sulfur cluster assembly (ISC) protein Iba57 executes a tetrahydrofolate-independent function in mitochondrial [4Fe-4S] protein maturation.
J.Biol.Chem., 298, 2022
|
|
7Z6K
 
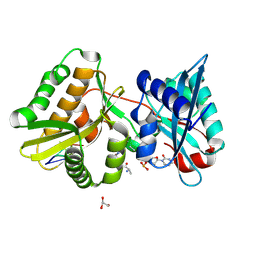 | |
7Z2I
 
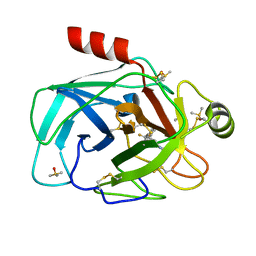 | | TRYPSIN (BOVINE) COMPLEXED WITH compound 4 | | Descriptor: | 5-[[3-(trifluoromethyl)phenyl]methyl]-1,4,6,7-tetrahydroimidazo[4,5-c]pyridine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schiering, N, Dalvit, C, Vulpetti, A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Efficient Screening of Target-Specific Selected Compounds in Mixtures by 19 F NMR Binding Assay with Predicted 19 F NMR Chemical Shifts.
Chemmedchem, 17, 2022
|
|
7Z25
 
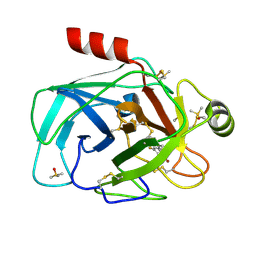 | | TRYPSIN (BOVINE) COMPLEXED WITH compound 12 | | Descriptor: | 2-(5-bromanyl-7-fluoranyl-2-methyl-1H-indol-3-yl)ethanamine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schiering, N, Dalvit, C, Vulpetti, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Efficient Screening of Target-Specific Selected Compounds in Mixtures by 19 F NMR Binding Assay with Predicted 19 F NMR Chemical Shifts.
Chemmedchem, 17, 2022
|
|
7Z5Z
 
 | |
7Z6A
 
 | |
7Z5Y
 
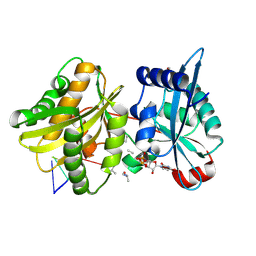 | |
7Z36
 
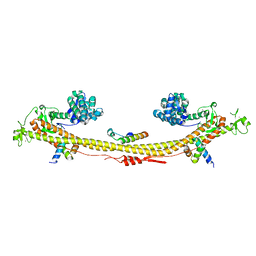 | | Crystal structure of the KAP1 tripartite motif in complex with the ZNF93 KRAB domain | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta,Isoform 2 of Transcription intermediary factor 1-beta, SMARCAD1 CUE1 domain, ZINC ION, ... | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional mapping of the KRAB-KAP1 repressor complex.
Embo J., 41, 2022
|
|
7Z7W
 
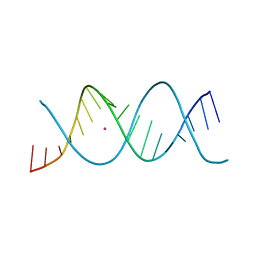 | | REP-related Chom18 variant with double GC base pairing | | Descriptor: | Chom18-GC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7Z
 
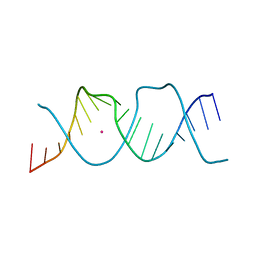 | | REP-related Chom18 variant with double TA base pair | | Descriptor: | Chom18-TA DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z82
 
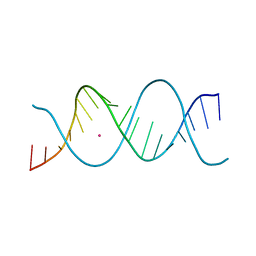 | | REP-related Chom18 variant with double AG mismatch | | Descriptor: | Chom18-AG DNA, STRONTIUM ION | | Authors: | Svoboda, J, Kolenko, P, Berdar, D, Schneider, B. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7K
 
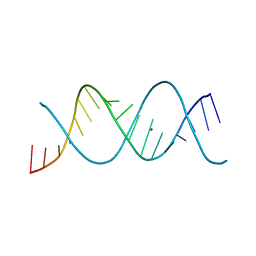 | | REP-related Chom18 variant with double AT base pairing | | Descriptor: | Chom18-AT DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
